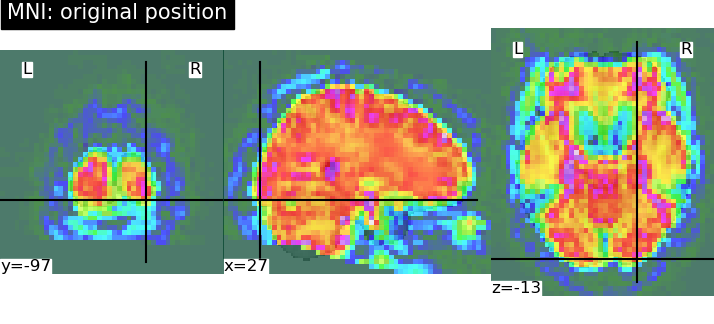
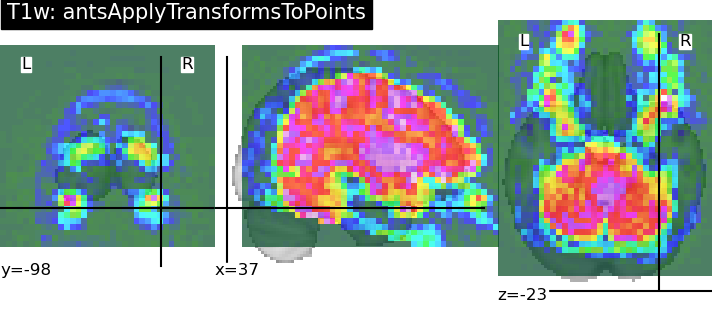
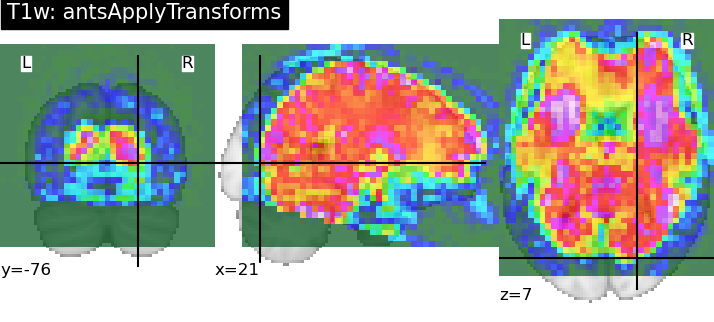
Hi there,
I’m new to using .h5 files, which fmriprep uses to store (composite) transforms. I have been able to successfully propagate labels from an individual to the template space:
antsApplyTransforms -d 3 --float 1 --verbose 1 -i sub-0086_T1w_label-aseg_roi.nii.gz -o output/sub-0086_T1w_space-MNI152NLin2009cAsym_label-aseg_roi.nii.gz -r MNI2009c_T1w.nii.gz -t sub-0086_T1w_target-MNI152NLin2009cAsym_warp.h5 -n NearestNeighbor
(Hypothetically) if I had the individual transform files (sub-0086_T1w_target-MNI152NLin2009cAsym_warp.nii.gz and sub-0086_T1w_target-MNI152NLin2009cAsym_affine.mat), I could have gotten the same result with the following call:
antsApplyTransforms -d 3 --float 1 --verbose 1 -i sub-0086_T1w_label-aseg_roi.nii.gz -o output/sub-0086_T1w_space-MNI152NLin2009cAsym_label-aseg_roi.nii.gz -r MNI2009c_T1w.nii.gz -t sub-0086_T1w_target-MNI152NLin2009cAsym_warp.nii.gz -t sub-0086_T1w_target-MNI152NLin2009cAsym_affine.mat -n NearestNeighbor
I have a particular use case where having these transformations separated would be beneficial; specifically, because I want to propagate some 3D points (e.g. anterior and posterior commissure) using antsApplyTransformsToPoints:
antsApplyTransformsToPoints -d 3 -i sub-0086_T1w_label-ACPC_points.csv -o output/sub-0086_T1w_space-MNI152NLin2009cAsym_label-ACPC_points.csv -t [sub-0086_T1w_target-MNI152NLin2009cAsym_affine.mat,1] -t sub-0086_T1w_target-MNI152NLin2009cAsym_inversewarp.nii.gz
Note that the ordering of linear and nonlinear transformations is reversed for point transformation, and also requires the inverse deformation field as input. For what it’s worth I tried inputting both forward and backward composite transforms derived from fmriprep and the coordinates were nowhere close to the true location (MNI2009c_T1w_label-ACPC_points.csv)
This is somewhat related to a few previous posts here and on the ants forum:
- FMRIPREP ANTS .h5 composite transform file decomposition? How exactly the .h5 decomposition was performed is not described and the focus of discussion was more on capturing the rigid component from the linear transformation matrix. @ChenChiaLan
- FMRIPREP: inverse t1_to_mni warp describing modifications to include both forward and inverse .h5 files.
-
https://sourceforge.net/p/advants/discussion/840260/thread/ff4587ef/?limit=25#c007 This covers the usage of
antsApplyTransformsToPoints
Any thoughts on how to resolve this? I’ve provided all the files as a .tar.gz link for anyone able to take a closer look.
-jon
–
Also, hope to see some of you at the INCF Montreal meeting next week!