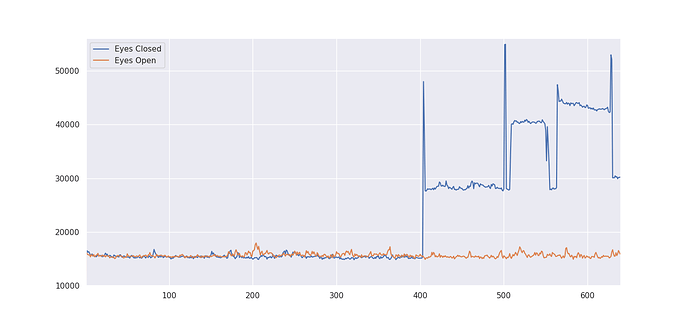
Hi all. Wanted to see if anyone else had run into this before.
I used AFNI’s 3dTo1D to calculate the euclidean norm, expecting it to be RELATIVELY similar to FD. However, the values I’ve gotten for the output are EXTREMELY high.
Range is from
This is SMS data, so I’m aware it should be notch filtered prior to any nuisance regression, but I am not doing so because what I’m examining is the relationship of the motion/respiration to independent components.
I’d like to know if I’m reading this right, because it looks like I have pretty large motion differences between my two conditions.
Has anyone encountered this before? Or is this a massive scaling SNAFU on my part?
These are the commands I used:
3dTto1D -input sub-${subjectID}/Session1/func/closed/sub-${subjectID}_Session1_task-restclosed_bold.nii.gz -method enorm -mask sub-${subjectID}/Session1/func/closed/Masked_Skullstripped_SBref_sub-${subjectID}_Session1_task-restclosed_bold.nii.gz -prefix $motiondir/sub-${subjectID}_ENORM.1D
1dplot -volreg -sepscl -jpg $motiondir/sub-${subjectID}_ENORM.jpg $motiondir/sub-${subjectID}_ENORM.1D
Thanks in advance.