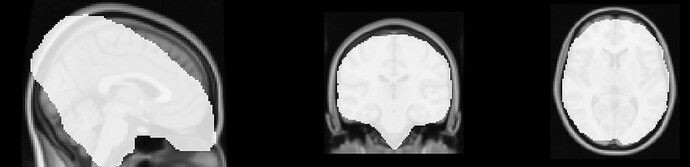
Hello,
The registration of my functional images to MNI space has failed, particularly in the sagittal (x) plane. The functional images have this stretching effect for all participants, for all runs, for the functional data only.
Here is an image of sub-*_ses-*_task-*_run-*_space-MNI52Lin2009cAsym_res-2_desc-brain_mask overlaid onto the MNI brain. Hopefully this demonstrates the issue.
Spatial normalisation of the anatomical images to MNI space is fine however. This is confirmed by in the logs, by looking at the clear correspondence between the reference T1w and MNI image.
What’s confusing, is that the logs did not indicate to me that this had occurred - everything looks okay here. But when viewing the spatial correspondence between the functional data (for all subjects, all tasks, all runs, all sessions) and MNI image, it’s obvious something has gone wrong.
To clarify, I used the following code when running fmriprep. Pretty standard stuff:
--output-spaces MNI152NLin2009cAsym:res-2 fsLR:den-32k
I’m really struggling to understand where this distortion has come from. If anyone has any ideas please let me know. I’m happy to provide more information/screenshots if needed.
Thank you very much,
Will