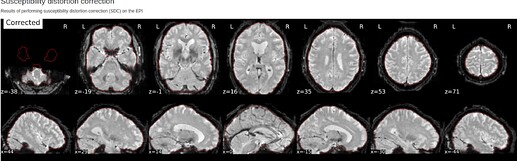
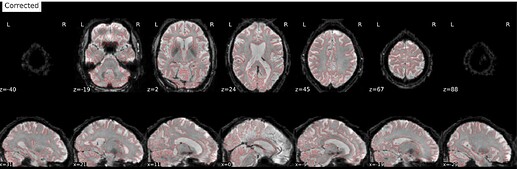
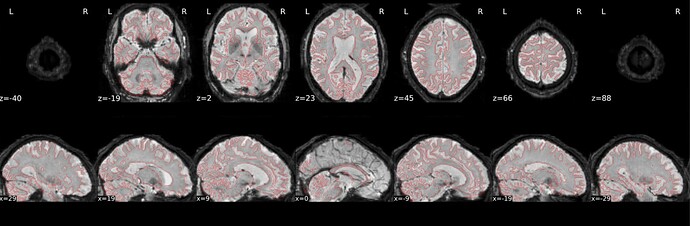
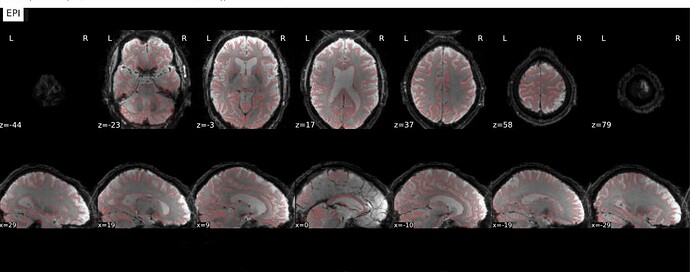
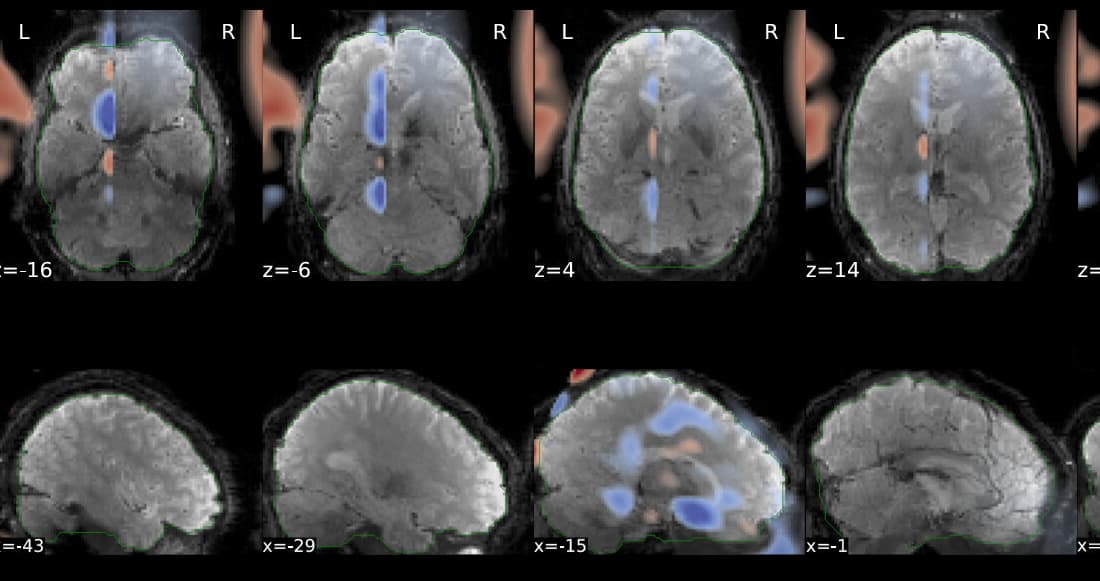
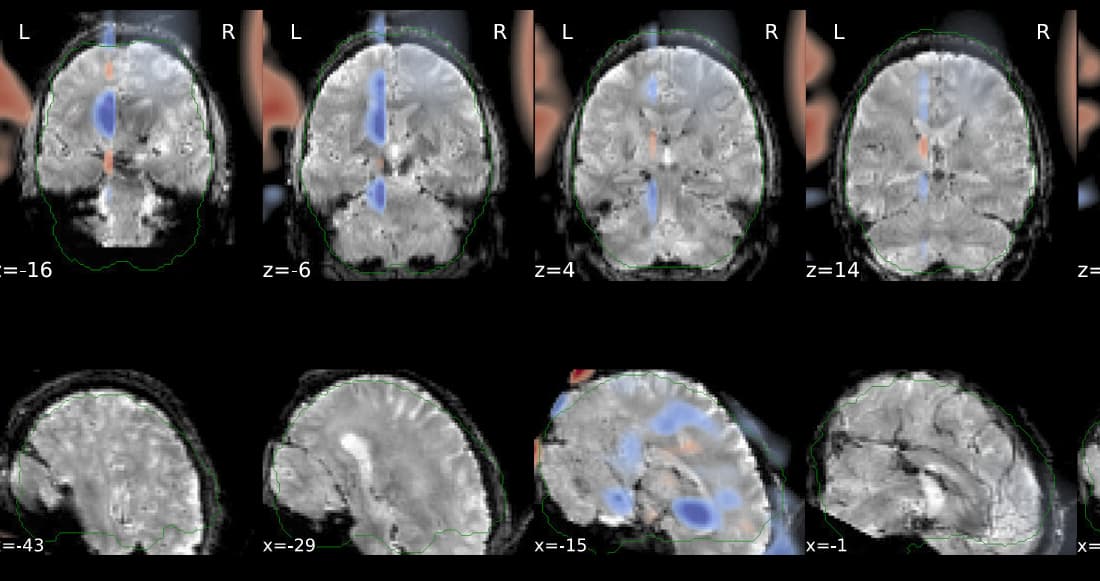
Hi @Steven,
here is the error report:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node syn.
Cmdline:
antsRegistration --collapse-output-transforms 1 --dimensionality 3 --initialize-transforms-per-stage 0 --interpolation Linear --output fmap_syn --transform SyN[ 0.8, 6.0, 3.0 ] --metric Mattes[ /scratch/fmriprep_23_2_wf/sub_pain36_wf/syn_preprocessing_auto_00001/anat2epi/clipped_noise_corrected_trans.nii.gz, /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/clip_epi/clipped.nii.gz, 0.5, 48, Random, 0.8 ] --metric Mattes[ /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/lap_anat_norm/clipped_noise_corrected_trans_maths_norm.nii.gz, /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/lap_epi_norm/clipped_maths_norm.nii.gz, 0.5, 48, Random, 0.8 ] --convergence [ 200x100, 1e-06, 5 ] --smoothing-sigmas 2.0x0.0vox --shrink-factors 1x1 --use-histogram-matching 1 --restrict-deformation 0.1x1.0x0.1 --masks [ /scratch/fmriprep_23_2_wf/sub_pain36_wf/syn_preprocessing_auto_00001/mask2epi/sub-pain36_desc-brain_mask_trans_uint8.nii.gz, /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/epi_umask/clipped_mask_union.nii.gz ] --transform SyN[ 0.8, 2.0, 1.0 ] --metric Mattes[ /scratch/fmriprep_23_2_wf/sub_pain36_wf/syn_preprocessing_auto_00001/anat2epi/clipped_noise_corrected_trans.nii.gz, /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/clip_epi/clipped.nii.gz, 0.5, 48 ] --metric Mattes[ /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/lap_anat_norm/clipped_noise_corrected_trans_maths_norm.nii.gz, /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/lap_epi_norm/clipped_maths_norm.nii.gz, 0.5, 48 ] --convergence [ 10, 1e-08, 2 ] --smoothing-sigmas 0.0vox --shrink-factors 1 --use-histogram-matching 1 --restrict-deformation 0.1x1.0x0.1 --masks [ /scratch/fmriprep_23_2_wf/sub_pain36_wf/syn_preprocessing_auto_00001/mask2epi/sub-pain36_desc-brain_mask_trans_uint8.nii.gz, /scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/epi_umask/clipped_mask_union.nii.gz ] --winsorize-image-intensities [ 0.001, 0.998 ] --write-composite-transform 0
Stdout:
Stderr:
Killed
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 453, in aggregate_outputs
setattr(outputs, key, val)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_types.py", line 2699, in validate
return TraitListObject(self, object, name, value)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 582, in __init__
super().__init__(
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 213, in __init__
super().__init__(self.item_validator(item) for item in iterable)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 213, in
super().__init__(self.item_validator(item) for item in iterable)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 865, in _item_validator
return trait_validator(object, self.name, value)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: Each element of the 'reverse_forward_transforms' trait of a RegistrationOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/syn/fmap_syn0Warp.nii.gz' was specified.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 400, in run
outputs = self.aggregate_outputs(runtime)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 460, in aggregate_outputs
raise FileNotFoundError(msg)
FileNotFoundError: No such file or directory '['/scratch/fmriprep_23_2_wf/sub_pain36_wf/fmap_preproc_wf/wf_auto_00001/syn/fmap_syn0Warp.nii.gz']' for output 'reverse_forward_transforms' of a FixHeaderRegistration interface
I did not have the time to look in it in detail, so please excuse me if there is an obvious error.