Hi!
I have a question about the Datagrabber node and how to iterate over subjects and sessions and files at the same time.
So I have a workflow with 150 subjects. Each of the subjects have 2 sessions and every session has ADC, FLAIR, T2* and masking data and several (20 ROI-masks). I have a previous workflow where I successfully iterated over all subjects with corresponding sessions. Now a I wanna elaborate that workflow and iterate (for each subject and session) over a list of ROI-names.
The problem that I’ve got right now is ther “iteration order”. It seems like it iterates over all ROI-files primarily. So for each ROI it chechs for a subject and session. I want it to be the opposite of that. Hope I’m making myself clear.
Also, it seems that it finds my ROI’s, here’s the report from the datagrabber:
Execution Outputs
- ADC : /home/brain/Documents/data/iNPH/BIDS/bids_nii/sub-136/ses-1/dwi/sub-136_ses-1_adc.nii.gz
- FLAIR : /home/brain/Documents/data/iNPH/BIDS/bids_nii/sub-136/ses-1/anat/sub-136_ses-1_FLAIR.nii.gz
- ROI_file : /home/brain/Documents/ROI/sub-136_ses-1/caudsin.nii
- t2star_mean : /home/brain/Documents/iNPH_workflow_test/datasink/4D_proc/sub-136_ses-1/mean_signal_masked.nii.gz
Here’s what I got so far:
from nipype import DataGrabber
from nipype.interfaces.io import DataSink
dg = Node(DataGrabber(infields=['subject_id', 'session_id', 'ROI' ],
outfields=['ADC', 'FLAIR', 't2star_mean', 'mask_t2star', 'ROI_file']),
name='datagrabber')
dg.inputs.base_directory = base_dir
# Necessary default parameters
dg.inputs.template = '*'
dg.inputs.sort_filelist = True
dg.inputs.field_template = {'ADC': bids_dir.replace(base_dir, '') + '/sub-%s/ses-%s/dwi/*adc.nii.gz',
'FLAIR': bids_dir.replace(base_dir, '') + '/sub-%s/ses-%s/anat/*FLAIR.nii.gz',
't2star_mean': datasink_dir.replace(base_dir, '') + '/4D_proc/sub-%s_ses-%s/mean_signal_masked.nii.gz',
'ROI_file': roi_dir.replace(base_dir, '') + '/sub-%s_ses-%s/%s.nii'}
dg.inputs.template_args['ADC'] = [['subject_id', 'session_id']]
dg.inputs.template_args['FLAIR'] = [['subject_id', 'session_id']]
dg.inputs.template_args['t2star_mean'] = [['subject_id', 'session_id']]
dg.inputs.template_args['ROI_file'] = [['subject_id', 'session_id', 'ROI']]
ROI_list = ['caudsin', 'cebref' , 'hippdx']
layout = BIDSLayout(patient_dir, validate = False)
subject_list = layout.get_subjects()
ses_list =layout.get_sessions()
Here comes the node from where I iterate:
infosource = Node(IdentityInterface(fields=['subject_id', 'session_id', 'ROI']),
name="infosource")
infosource.iterables = [('subject_id', subject_list),
('session_id', ses_list),
('ROI', ROI_list)]
Here’s where I connect infosource to the datagrabber node (which is in the “wf_pat” workflow)
wf_stud = Workflow(name="studieproc", base_dir = output_dir)
wf_stud.connect([(infosource, wf_pat, [("subject_id", "datagrabber.subject_id"),
("session_id", "datagrabber.session_id"),
("ROI", "datagrabber.ROI")]),
])
wf_stud.run()
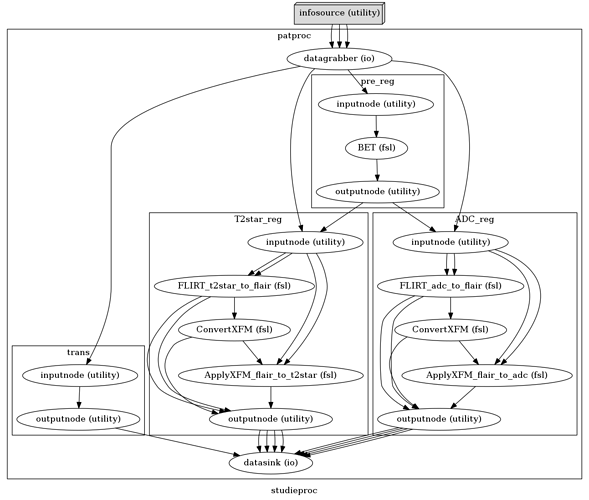
Here’s just an overview of the workflow. It’s in the “trans” workflow I wanna process the ROI’s. (I’m gonna transform them with the matrix gotten from the other two workflows (t2star_reg and ADC-reg)
Sincerely, Jesper