Summary of what happened:
BIDS layout in XCP-D doesn’t seem to recognize the T1w scan. I haven’t run the validator since I am pretty sure it will fail: the derivatives were selectively downloaded via NDA from ABCC. But as far as I understand xcp_d can still run, and the layout validator in XCP-D is disabled either way. The download included:
- T1w MNI152NLin2009cAsym
- BOLD MNI152NLin2009cAsym
- Confounds.
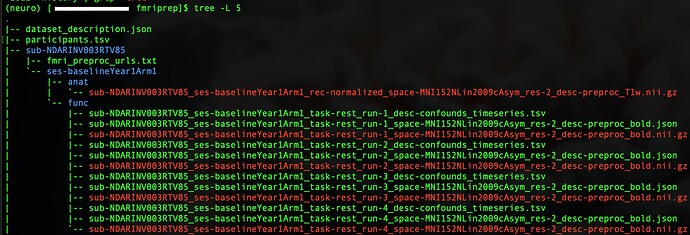
fMRIPrep directory tree:
|-- dataset_description.json
|-- participants.tsv
| |-- sub-NDARINVMENCHP62
| | `-- ses-baselineYear1Arm1
| | |-- anat
| | | `-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_space-MNI152NLin2009cAsym_res-2_desc-preproc_T1w.nii.gz
| | `-- func
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-1_desc-confounds_timeseries.tsv
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-1_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.json
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-1_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.nii.gz
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-2_desc-confounds_timeseries.tsv
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-2_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.json
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-2_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.nii.gz
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-3_desc-confounds_timeseries.tsv
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-3_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.json
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-3_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.nii.gz
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-4_desc-confounds_timeseries.tsv
| | |-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-4_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.json
| | `-- sub-NDARINVMENCHP62_ses-baselineYear1Arm1_task-rest_run-4_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.nii.gz
Command used:
singularity exec -B $MRIS -B $HOME -B $SIMG $SIMG/xcp_d-0.9.0.simg /usr/local/miniconda/bin/xcp_d \
$MRIS \
$XCPD_OUT \
participant \
--mode abcd \
--input-type fmriprep \
--motion-filter-type none \
--participant_label $sub \
--work-dir $XCPD_work \
-p 27P \
-t rest \
--file-format nifti \
-f 0.3 \
--lower-bpf 0.009 \
--upper-bpf 0.08 \
--fs-license-file $FS_LICENSE \
--stop-on-first-crash \
--warp-surfaces-native2std n \
--atlases '4S456Parcels' \
-vvv
Version:
0.9.0
Environment:
Apptainer 1.2.5-1.el8
Error traceback:
File "/usr/local/miniconda/lib/python3.10/multiprocessing/process.py", line 314, in _bootstrap
self.run()
File "/usr/local/miniconda/lib/python3.10/multiprocessing/process.py", line 108, in run
self._target(*self._args, **self._kwargs)
File "/usr/local/miniconda/lib/python3.10/site-packages/xcp_d/cli/workflow.py", line 99, in build_workflow
retval["workflow"] = init_xcpd_wf()
File "/usr/local/miniconda/lib/python3.10/site-packages/xcp_d/workflows/base.py", line 78, in init_xcpd_wf
single_subject_wf = init_single_subject_wf(subject_id)
File "/usr/local/miniconda/lib/python3.10/site-packages/xcp_d/workflows/base.py", line 124, in init_single_subject_wf
subj_data = collect_data(
File "/usr/local/miniconda/lib/python3.10/site-packages/xcp_d/utils/bids.py", line 268, in collect_data
raise FileNotFoundError("No T1w or T2w files found.")
FileNotFoundError: No T1w or T2w files found.