Hi. I have a set of mri data with the shape (121,145,121) and voxel size 1.5mm × 1.5mm × 1.5mm. I’ve applied searchlight on it and I have an array of scores with the shape (121,145,121). I want to calculate the average of scores in each region of the AAL atlas. But I don’t know how to find voxels corresponding to each region to calculate their average. Would you please help me?
Thank you in advance
Hi @Sara,
Is your data in the same space as the AAL atlas? If so, consider using nilearn.maskers.NiftiLabelsMasker.
Best,
Steven
I don’t even know how to get an AAL atlas. Would you please explain more?
You can fetch the atlas also through Nilearn: nilearn.datasets.fetch_atlas_aal - Nilearn . I believe it is in MNI space, so if your searchlight analysis is in MNI you should be all set.
Best,
Steven
Yes my data is in MNI space, too. So isn’t there any problem that my data’s voxel sizes are 1.5×1.5×1.5?
Shouldn’t be a problem!
Thanks a lot for your guidance.
Would you please explain more about how exactly I can use this to find each voxels region?
If you fit the label masker made by AAL atlas labels on your searchlight image, it returns the average searchlight result per AAL label.
The AAL atlas is available through a few places including distribution within MRIcron. It’s very similar to the macrolabel atlas for the N27 dataset that is distributed with the cytoarchitectonic atlases from Eickhoff-Zilles and now the JuBrain anatomy toolbox. The macrolabel atlas is provided with AFNI. For integration with AFNI and more about extraction, there is the discussion from a few years ago:
Would you please give me the exact code of how to do this? I’m new to neuroimaging and I’m so confused now.
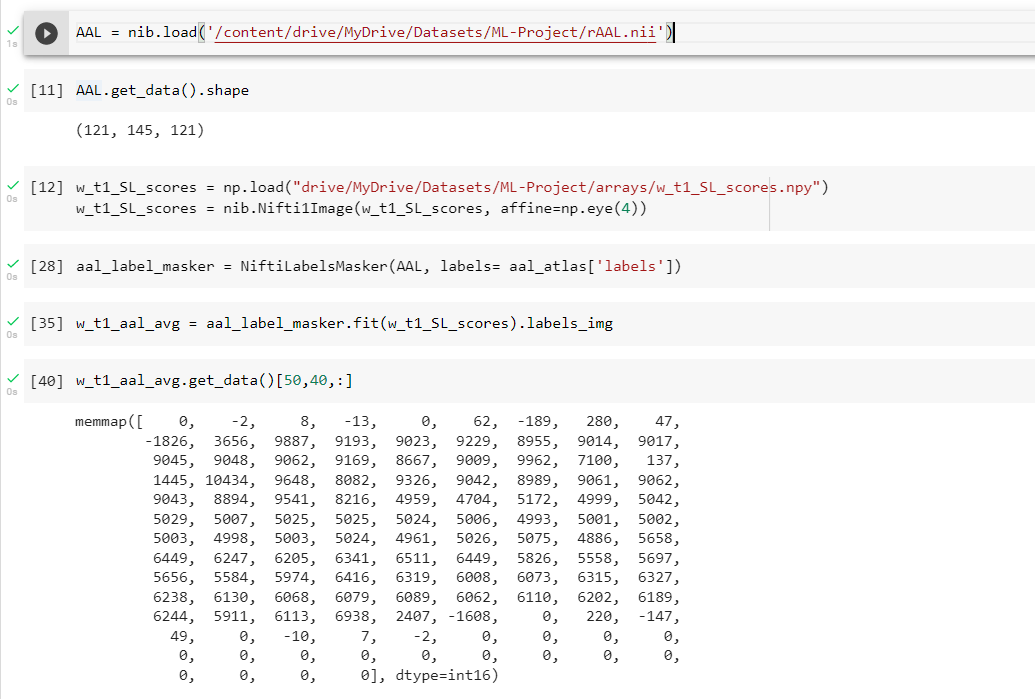
I downloaded AAL atlas from fetch_aal_atlas and with SPM tools I converted this atlas to my data’s shape (121,145,121). Then I did this
What I have got? What are these numbers?
I want to know exactly each voxel belongs to what region. So then I can calculate the average score of each regions’ voxels’ searchlight’s score. Then I want to pick the voxels in the region with the highest average score and apply the SVM on them. So I need to know each voxel belongs to what region.
import nilearn.datasets, nilearn.maskers, nilearn.image
# Load searchlight results
searchlight_img = nilearn.image.load_img('/PATH/TO/SEARCHLIGHT/IMAGE.nii.gz')
# Load the AAL atlas
aal = nilearn.datasets.fetch_atlas_aal()
aal_img = aal['maps']
aal_labels = aal['labels']
# Create the masker
aal_masker = nilearn.maskers.NiftiLabelsMasker(aal_img, labels=aal_labels)
# Fit the masker
average_searchlight_vals = aal_masker.fit_transform(imgs=searchlight_img)
The output is an array the size of your labels array, where the nth element is the average searchlight coefficient in the nth label of the AAL atlas.
For you other point about finding which voxels are in a given region, you could start by resampling the AAL atlas image to dimensions of your searchlight image
aal_resampled = nilearn.image.resample_img(aal_img, target_affine=XX, target_shape=XX, interpolation='nearest') where target_affine and target_shape would come from your searchlight image.
Then you can make a mask for a given region with something like
aal_img_loaded = nilearn.image.load_img(aal_img)
aal_data_array = aal_img_loaded.get_fdata()
mask_region = (aal_data_array==2001) # 2001 is one of the label values of AAL
searchlight_data_array = searchlight_img.get_fdata()
searchlight_data_in_mask = searchlight_data_array[mask_region]
Thanks @Steven . Please I have a follow up question
From your first response, is the output from the “average_searchlight_vals”, the average intensity of each ROI in the AAL atlas or the average volume of each ROI? If the output was the average intensity, how do I get the total volume of each ROI using this atlas?
From the documentation, I noticed it returns region_signals, but I was wondering what the signal is ? I am applying it on a 3D S-MRI image, with the goal of extracting the total volume of each ROI.
Thanks in Advance.
Hi @dlaplace01 and welcome to neurostars!
It is returning the average intensity of the input image in each ROI of the AAL atlas.
To get volume you can just count the number of voxels in the label image that correspond to a given ROI (look at the code about ROI masking in my earlier post above) and multiply it by the volume per voxel in the atlas.
Best,
Steven
Thanks Steven.
But here my problem is when I resample AAL to the dimensions of my data, there will be 23 different numbers in it. And therefore I can’t calculate the average score of the actual regions of AAL atlas.
Can you show with code outputs how you are getting this number of 23? It doesn’t make sense that you would lose regions because the AAL atlas from Nilearn is 2mm isotopic. So upsampling the atlas should not result in losing data.
aal_atlas = fetch_atlas_aal()
AAL = nib.load(aal_atlas['maps'])
aal_data = AAL.get_data()
vals = []
for i in range(91):
for j in range(109):
for z in range(91):
if aal_data[i,j,z] not in vals:
vals.append(aal_data[i,j,z])
len(vals)
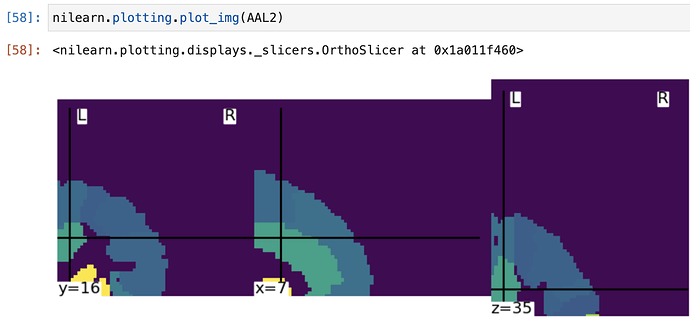
AAL2 = resample_img(AAL, target_affine = np.eye(4), target_shape = (121,145,121),
interpolation = 'nearest')
aal2_data = AAL2.get_data()
vals2 = []
for i in range(121):
for j in range(145):
for z in range(121):
if aal2_data[i,j,z] not in vals2:
vals2.append(aal2_data[i,j,z])
len(vals2)
The length of vals2 = 23
A few things:
- Instead of looping over all the directions, you can just use
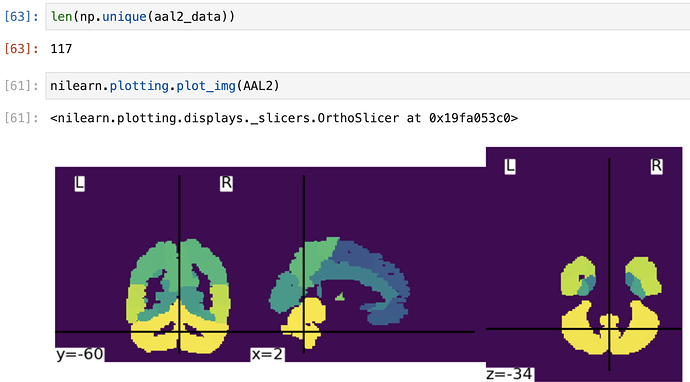
numpy.unique(array)to get unique values of an array. - Your affine is incorrect in the image resampling, as illustrated below when I plot the resampled image.
Simply changing the affine toAAL.affinefixes it!
Best,
Steven