Hello,
I’m wondering about the validity of my current approach. I have voxel-level betas for each stimulus activation for 30 separate sessions in one subject. There are two conditions I’m interested in for each Session:
A (called ‘Session’ in design matrix)- [0 or 1]
B (called ‘Face’ in design matrix)- [0 or 1]
I’m hoping to perform an interaction analysis (AxB) between both variables for each session, and then perform second level analysis across all sessions. My first-level design (for each session) is below:
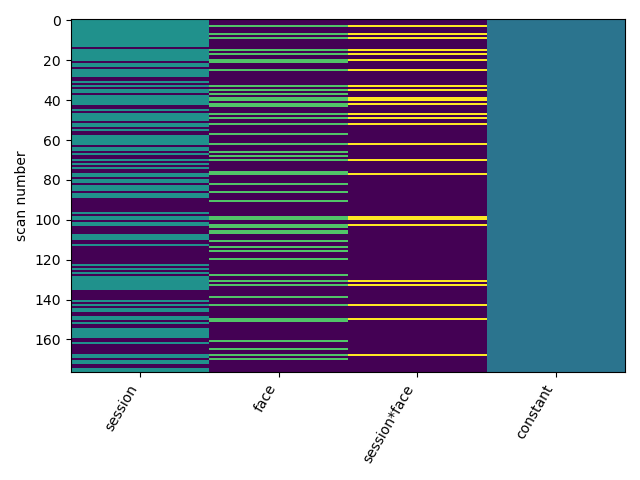
My first-level contrast was specified as [0 0 1 0]. I then performed a one-sample t-test of effect sizes across all Sessions.
Is this a feasible way to assess the interaction of variables per Session, and conglomerate the results? I’m confused as to what a positive and negative effect could be associated with here exactly. I could perform a post-hoc test of certain clusters to further analyze.
Thanks in advance.
“s this a feasible way to assess the interaction of variables per Session, and conglomerate the results?”
Yes, I would do so.
“I’m confused as to what a positive and negative effect could be associated with here exactly”
Maybe the interpretation looks a bit challenging because the main effects are not centered, i.e. not orthogonal to the constant vector, but actually the model does what you expect it does.
2 Likes
Thank you for your feedback. It is very appreciated!
Hi @bthirion , I’ve got a quick follow-up question.
If I’m interested in seed-connectivity based AxB interaction–is it possible to change my design matrix columns to:
SEEDxA, SEEDxB,[(SEEDxA)x(SEEDxB)], Constant
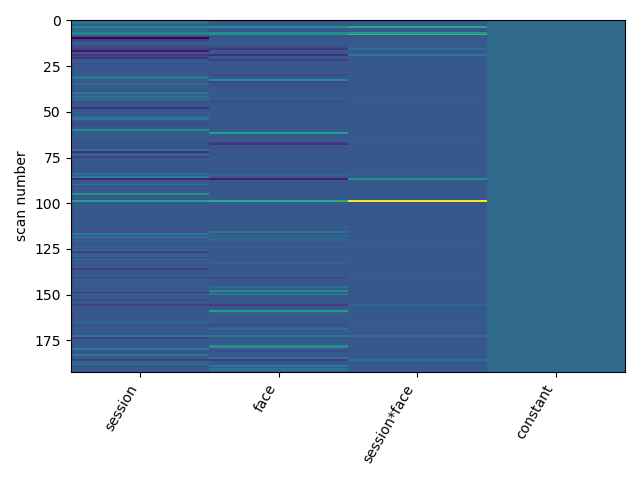
to run this analysis? Does that make sense?
Thanks,
Tom
I’m I think I would put a SEED regressor on top of the interaction regressors.
Intuitively SEEDxAxB makes more sense to me than [(SEEDxA)x(SEEDxB)].
On thing I’m realizing: SEED corresponds to BOLD signal, i.e. delayed through the hrf response. So A and B should also be time onsets convolved with the hrf, otherwise, you may be multiplying apples and oranges.
Best,
Hi @bthirion , I have a general question about second-level analysis if you have time to answer. I have been using the ‘effect_size’ output from first-level compute_contrasts as my second-level input, is this indeed correct? Or should I be using the ‘stat’ output from my first-level compute_contrasts? Is this depended on a certain research question?
In theory you should use the effect size, because this allows you to interpret the final statistics as BOLD signal change, making the final statistics physically grounded.
Now, there is nothing wrong with using the stat instead. The advantage of stat is that its is better normalized across individuals (the effect size is not necessarily directly comparable). I usually do that.
2 Likes

