Hello!
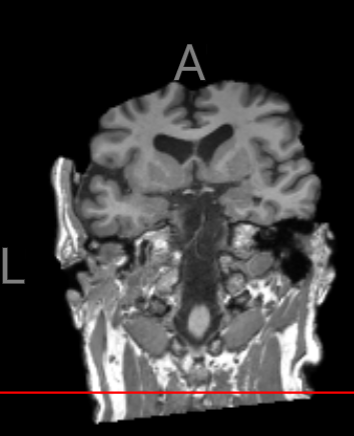
After applying some preprocessing steps to my ADNI data, the samples are mostly clean, but there are still some samples that contain neck artifacts.
How can I address this issue? Is there a specific step or set of parameters I could use to make adjustments to all the samples? Manually inspecting 944 samples would be quite challenging. samples would be quite challenging.
Please refer to my prepocessing bash below:
#!/bin/bash
apply_bias_correction() {
local input_dir=$1
local output_dir=$2
# Count total number of files
total_files=$(find "$input_dir" -name "*_normalized.nii.gz" | wc -l)
current_file=0
for file in "$input_dir"/*_normalized.nii.gz; do
if [ -f "$file" ]; then
filename=$(basename "$file" _normalized.nii.gz)
output_file="$output_dir/${filename}_bias_corrected.nii.gz"
# Skip if already processed
if [ -f "$output_file" ]; then
echo "Skipping already processed file: $output_file"
continue
fi
((current_file++))
# Calculate progress percentage
progress=$((current_file * 100 / total_files))
# Print progress bar
printf "\rProgress: [%-50s] %d%%" $(printf "#%.0s" $(seq 1 $((progress/2)))) $progress
# Apply bias field correction using FSL's FAST
fast -t 1 -n 3 -H 0.1 -I 4 -l 20.0 -B -o "$output_dir/${filename}_bias_corrected" "$file" > /dev/null 2>&1
# Move and rename the bias-corrected image
mv "$output_dir/${filename}_bias_corrected_restore.nii.gz" "$output_file"
fi
done
# Print newline after progress bar
echo
}
echo "Processing training data..."
apply_bias_correction "$train_dir" "$output_train_dir"
echo "Processing testing data..."
apply_bias_correction "$test_dir" "$output_test_dir"
echo "Bias field correction complete for all files."