Hello everyone,
I am trying to register a brain-extracted T2 FLAIR image into MNI space using FLIRT.
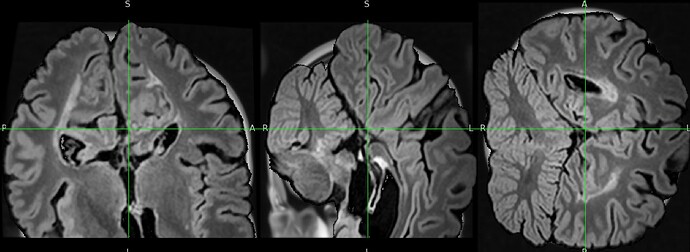
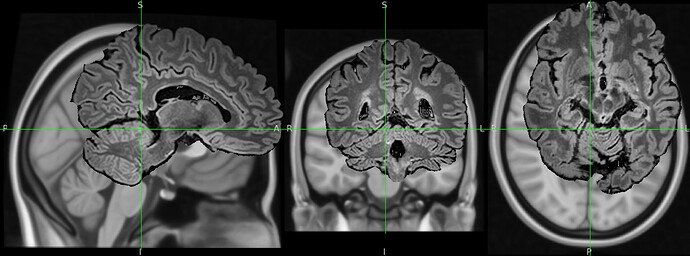
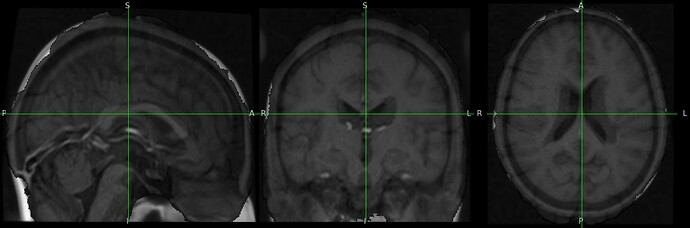
The output image is totally distorded and cropped of and i don’t understand why.
The fslinfo of my file :
filename FLAIR_2_Brain.nii.gz
sizeof_hdr 348
data_type FLOAT32
dim0 3
dim1 380
dim2 480
dim3 340
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 16
nbyper 4
bitpix 32
pixdim0 1.000000
pixdim1 0.500000
pixdim2 0.500000
pixdim3 0.500000
pixdim4 8.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 0.499851 -0.000183 -0.012216 -103.352325
qto_xyz:2 0.001787 0.495659 0.065720 -112.136017
qto_xyz:3 0.012086 -0.065744 0.495511 -42.285835
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Left-to-Right
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 0.499847 -0.000183 -0.012216 -103.352325
sto_xyz:2 0.001787 0.495659 0.065720 -112.136017
sto_xyz:3 0.012086 -0.065744 0.495511 -42.285835
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Left-to-Right
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip 2203.12-dirty 2024-02-01T16:17:47+00:00
aux_file
The template file :
filename MNI152_T1_0.5mm.nii.gz
sizeof_hdr 348
data_type UINT8
dim0 3
dim1 364
dim2 436
dim3 364
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 2
nbyper 1
bitpix 8
pixdim0 -1.000000
pixdim1 0.500000
pixdim2 0.500000
pixdim3 0.500000
pixdim4 1.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 250.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name MNI_152
qform_code 4
qto_xyz:1 -0.500000 0.000000 -0.000000 90.000000
qto_xyz:2 0.000000 0.500000 -0.000000 -126.000000
qto_xyz:3 0.000000 0.000000 0.500000 -72.000000
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name MNI_152
sform_code 4
sto_xyz:1 -0.500000 0.000000 0.000000 90.000000
sto_xyz:2 0.000000 0.500000 0.000000 -126.000000
sto_xyz:3 0.000000 0.000000 0.500000 -72.000000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip FSL5.0
aux_file
I am using this flirt line, via the GUI :
flirt -in /Users/.../FLAIR_2_brain.nii.gz -ref /Users/.../MNI152_T1_0.5mm.nii.gz -out /Users/.../FLAIR_2_registred.nii.gz -omat /Users/.../FLAIR_2_registred.mat -bins 256 -cost corratio -searchrx -90 90 -searchry -90 90 -searchrz -90 90 -dof 12 -interp trilinear
The result totally cropped and distorded and i can’t figure out what went wrong:
Could you help me understand the issue ?
Thank you so much !