Hi all,
I have a nifti file that is used by some code I am developing. I can open and use the nifti file locally. I can push the .nii file to github, where my repository for the code is. It is about 4 MB in size. However, when I download the file or clone the repo, when I try to open the file, I get a floating point overflow error in MRIcron, and I am unable to use the file in my code.
Has anyone dealt with something like this before? I’m not sure if there are special procedures I need to take with large files in github or how to approach the problem.
Oh, MRIcroGL gives me a more specific error (Error -5 during UnCompressMem process). Does this have something to do with the .gz compression?
Here is the repository. The files that I’m having trouble with are:
icbm152_flair_brain.nii.gz
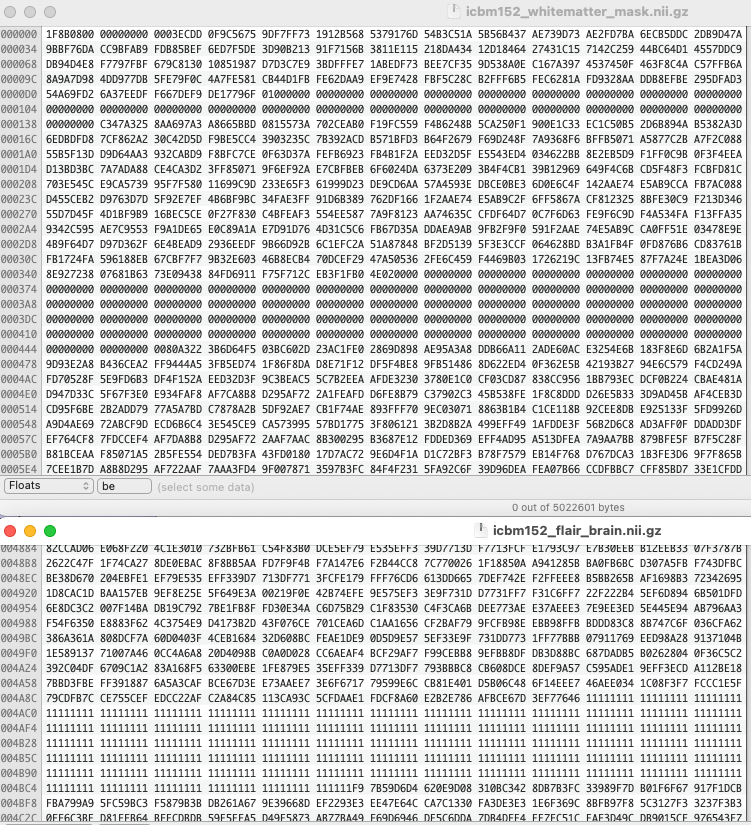
icbm152_whitematter_mask.nii.gz
The gz files are clearly corrupted.
lesion-mapper > gzip -k -d icbm152_flair_brain.nii.gz
gzip: invalid compressed data--crc error
gzip: icbm152_flair_brain.nii.gz: uncompress failed
lesion-mapper > gzip -V
Apple gzip 321.100.11
A hex editor shows long runs of 1111... in one file and 00000... in the other. This is pretty improbable - one of the major strategies for compression is to remove predictable repeating patterns. Not sure what happened here, this does not look like the classic corruption of binary data by a ftp transfer in the default ASCII mode.
You might want to check how your are uploading your file to github. If you use a macOS computer, I highly recommend the free and open source GitUp.
1 Like
Thank you so much for honing me in on the upload process. The upload was not working. I wasn’t planning on ever changing these image files, but I updated to git-lfs and it seemed to upload without corrupting the file.