Hi there,
First post here on Neurostars.org. Should have joined this a long time ago but better late than never.
I am running an analysis on a subset of the original OASIS database (anatomical only). I’ve been using fmriprep 1.1.1 on a subset of 30 subjects using the following command on a slurm cluster:
singularity run -B /localscratch:/localscratch -e -B /cvmfs:/cvmfs -B /project:/project -B /scratch:/scratch /opt/singularity/bids-apps/poldracklab_fmriprep_1.1.1.img /home/user/study/PHASE2/input_data/OAS1_bids /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1 participant --participant_label $subj --nthreads 8 --n_cpus 8 --omp-nthreads 8 --mem_mb 32000 --debug -w /scratch/${USER}/${SLURM_JOB_ID}_${SLURM_ARRAY_TASK_ID} --fs-license-file /opt/freesurfer/.license --resource-monitor --notrack --anat-only
29 out of 30 completed successfully but one subject (sub-0180) failed (see bottom of message for the full output log).
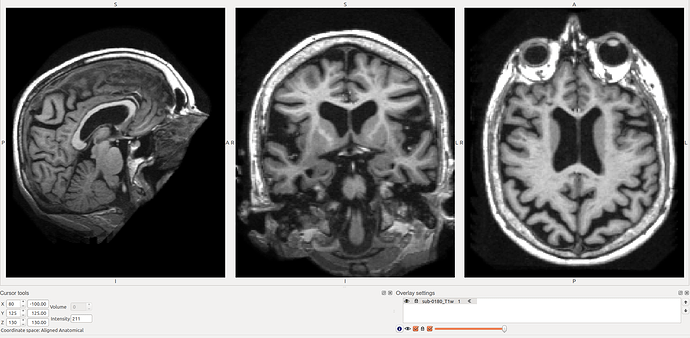
Here’s a screenshot of sub-0180. Ventricles large (not so unusual in this cohort) and the head/neck is perhaps more extended than most.
It appears to be failing on one of the freesurfer steps encapsulated in fmriprep. If I was running freesurfer on its own I would likely try out some of the strategies suggested in the output log. However, just wanted to ask here about whether there are some best practices, guidelines, or suggestions on how to successfully get an output without deviating too much from the bids-app workflow.
Appreciate your time, and thanks to @ChrisGorgolewski and colleagues for making this tool available!
Let me know if you need any more clarification.
Best regards,
jon
The error message is the following:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/plugins/multiproc.py", line 68, in run_node
result['result'] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py", line 480, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py", line 564, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py", line 644, in _run_command
result = self._interface.run(cwd=outdir)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/interfaces/base/core.py", line 521, in run
runtime = self._run_interface(runtime)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/interfaces/base/core.py", line 1044, in _run_interface
self.raise_exception(runtime)
File "/usr/local/miniconda/lib/python3.6/site-packages/nipype/interfaces/base/core.py", line 981, in raise_exception
).format(**runtime.dictcopy()))
RuntimeError: Command:
recon-all -autorecon1 -i /scratch/user/6427737_9/fmriprep_wf/single_subject_0180_wf/anat_preproc_wf/anat_template_wf/t1_conform/mapflow/_t1_conform0/sub-0180_T1w_ras.nii.gz -noskullstrip -openmp 8 -subjid sub-0180 -sd /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer
Standard output:
Subject Stamp: freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.1-f53a55a
Current Stamp: freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.1-f53a55a
INFO: SUBJECTS_DIR is /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer
Actual FREESURFER_HOME /opt/freesurfer
Linux node643 3.10.0-693.11.6.el7.x86_64 #1 SMP Thu Jan 4 01:06:37 UTC 2018 x86_64 x86_64 x86_64 GNU/Linux
'/opt/freesurfer/bin/recon-all' -> '/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/scripts/recon-all.local-copy'
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180
mri_convert /scratch/user/6427737_9/fmriprep_wf/single_subject_0180_wf/anat_preproc_wf/anat_template_wf/t1_conform/mapflow/_t1_conform0/sub-0180_T1w_ras.nii.gz /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig/001.mgz
mri_convert.bin /scratch/user/6427737_9/fmriprep_wf/single_subject_0180_wf/anat_preproc_wf/anat_template_wf/t1_conform/mapflow/_t1_conform0/sub-0180_T1w_ras.nii.gz /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig/001.mgz
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from /scratch/user/6427737_9/fmriprep_wf/single_subject_0180_wf/anat_preproc_wf/anat_template_wf/t1_conform/mapflow/_t1_conform0/sub-0180_T1w_ras.nii.gz...
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (1, 0, 0)
j_ras = (0, 1, 0)
k_ras = (0, 0, 1)
writing to /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig/001.mgz...
#--------------------------------------------
#@# MotionCor Wed Jul 18 00:08:16 UTC 2018
Found 1 runs
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig/001.mgz
Checking for (invalid) multi-frame inputs...
WARNING: only one run found. This is OK, but motion
correction cannot be performed on one run, so I'll
copy the run to rawavg and continue.
cp /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig/001.mgz /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/rawavg.mgz
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180
mri_convert /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/rawavg.mgz /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig.mgz --conform
mri_convert.bin /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/rawavg.mgz /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig.mgz --conform
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/rawavg.mgz...
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (1, 0, 0)
j_ras = (0, 1, 0)
k_ras = (0, 0, 1)
changing data type from float to uchar (noscale = 0)...
MRIchangeType: Building histogram
Reslicing using trilinear interpolation
writing to /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig.mgz...
mri_add_xform_to_header -c /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/transforms/talairach.xfm /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig.mgz /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/orig.mgz
INFO: extension is mgz
#--------------------------------------------
#@# Talairach Wed Jul 18 00:08:23 UTC 2018
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
mri_nu_correct.mni --no-rescale --i orig.mgz --o orig_nu.mgz --n 1 --proto-iters 1000 --distance 50
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
/opt/freesurfer/bin/mri_nu_correct.mni
--no-rescale --i orig.mgz --o orig_nu.mgz --n 1 --proto-iters 1000 --distance 50
nIters 1
$Id: mri_nu_correct.mni,v 1.27 2016/02/26 16:19:49 mreuter Exp $
Linux node643 3.10.0-693.11.6.el7.x86_64 #1 SMP Thu Jan 4 01:06:37 UTC 2018 x86_64 x86_64 x86_64 GNU/Linux
Wed Jul 18 00:08:23 UTC 2018
Program nu_correct, built from:
Package MNI N3, version 1.12.0, compiled by nicks@terrier (x86_64-unknown-linux-gnu) on 2015-06-19 at 01:25:34
/usr/bin/bc
tmpdir is ./tmp.mri_nu_correct.mni.29315
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
mri_convert orig.mgz ./tmp.mri_nu_correct.mni.29315/nu0.mnc -odt float
mri_convert.bin orig.mgz ./tmp.mri_nu_correct.mni.29315/nu0.mnc -odt float
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from orig.mgz...
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
changing data type from uchar to float (noscale = 0)...
writing to ./tmp.mri_nu_correct.mni.29315/nu0.mnc...
--------------------------------------------------------
Iteration 1 Wed Jul 18 00:08:25 UTC 2018
nu_correct -clobber ./tmp.mri_nu_correct.mni.29315/nu0.mnc ./tmp.mri_nu_correct.mni.29315/nu1.mnc -tmpdir ./tmp.mri_nu_correct.mni.29315/0/ -iterations 1000 -distance 50
[user@node643:/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/] [2018-07-18 00:08:25] running:
/opt/freesurfer/mni/bin/nu_estimate_np_and_em -parzen -log -sharpen 0.15 0.01 -iterations 1000 -stop 0.001 -shrink 4 -auto_mask -nonotify -b_spline 1.0e-7 -distance 50 -quiet -execute -clobber -nokeeptmp -tmpdir ./tmp.mri_nu_correct.mni.29315/0/ ./tmp.mri_nu_correct.mni.29315/nu0.mnc ./tmp.mri_nu_correct.mni.29315/nu1.imp
Processing:.................................................................Done
Processing:.................................................................Done
Processing:.................................................................Done
Number of iterations: 76
CV of field change: 0.000995587
mri_convert ./tmp.mri_nu_correct.mni.29315/nu1.mnc orig_nu.mgz --like orig.mgz --conform
mri_convert.bin ./tmp.mri_nu_correct.mni.29315/nu1.mnc orig_nu.mgz --like orig.mgz --conform
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from ./tmp.mri_nu_correct.mni.29315/nu1.mnc...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
INFO: transform src into the like-volume: orig.mgz
changing data type from float to uchar (noscale = 0)...
MRIchangeType: Building histogram
writing to orig_nu.mgz...
Wed Jul 18 00:09:57 UTC 2018
mri_nu_correct.mni done
talairach_avi --i orig_nu.mgz --xfm transforms/talairach.auto.xfm
talairach_avi log file is transforms/talairach_avi.log...
Started at Wed Jul 18 00:09:57 UTC 2018
Ended at Wed Jul 18 00:10:21 UTC 2018
talairach_avi done
cp transforms/talairach.auto.xfm transforms/talairach.xfm
#--------------------------------------------
#@# Talairach Failure Detection Wed Jul 18 00:10:23 UTC 2018
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
talairach_afd -T 0.005 -xfm transforms/talairach.xfm
ERROR: talairach_afd: Talairach Transform: transforms/talairach.xfm ***FAILED*** (p=0.0697, pval=0.0034 < threshold=0.0050)
Manual Talairach alignment may be necessary, or
include the -notal-check flag to skip this test,
making sure the -notal-check flag follows -all
or -autorecon1 in the command string.
See:
http://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/Talairach
INFO: Retrying Talairach align using 3T-based atlas...
#--------------------------------------------
#@# Talairach Wed Jul 18 00:10:24 UTC 2018
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
mri_nu_correct.mni --no-rescale --i orig.mgz --o orig_nu.mgz --n 1 --proto-iters 1000 --distance 50
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
/opt/freesurfer/bin/mri_nu_correct.mni
--no-rescale --i orig.mgz --o orig_nu.mgz --n 1 --proto-iters 1000 --distance 50
nIters 1
$Id: mri_nu_correct.mni,v 1.27 2016/02/26 16:19:49 mreuter Exp $
Linux node643 3.10.0-693.11.6.el7.x86_64 #1 SMP Thu Jan 4 01:06:37 UTC 2018 x86_64 x86_64 x86_64 GNU/Linux
Wed Jul 18 00:10:24 UTC 2018
Program nu_correct, built from:
Package MNI N3, version 1.12.0, compiled by nicks@terrier (x86_64-unknown-linux-gnu) on 2015-06-19 at 01:25:34
/usr/bin/bc
tmpdir is ./tmp.mri_nu_correct.mni.32667
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
mri_convert orig.mgz ./tmp.mri_nu_correct.mni.32667/nu0.mnc -odt float
mri_convert.bin orig.mgz ./tmp.mri_nu_correct.mni.32667/nu0.mnc -odt float
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from orig.mgz...
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
changing data type from uchar to float (noscale = 0)...
writing to ./tmp.mri_nu_correct.mni.32667/nu0.mnc...
--------------------------------------------------------
Iteration 1 Wed Jul 18 00:10:25 UTC 2018
nu_correct -clobber ./tmp.mri_nu_correct.mni.32667/nu0.mnc ./tmp.mri_nu_correct.mni.32667/nu1.mnc -tmpdir ./tmp.mri_nu_correct.mni.32667/0/ -iterations 1000 -distance 50
[user@node643:/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/] [2018-07-18 00:10:25] running:
/opt/freesurfer/mni/bin/nu_estimate_np_and_em -parzen -log -sharpen 0.15 0.01 -iterations 1000 -stop 0.001 -shrink 4 -auto_mask -nonotify -b_spline 1.0e-7 -distance 50 -quiet -execute -clobber -nokeeptmp -tmpdir ./tmp.mri_nu_correct.mni.32667/0/ ./tmp.mri_nu_correct.mni.32667/nu0.mnc ./tmp.mri_nu_correct.mni.32667/nu1.imp
Processing:.................................................................Done
Processing:.................................................................Done
Processing:.................................................................Done
Number of iterations: 76
CV of field change: 0.000995587
mri_convert ./tmp.mri_nu_correct.mni.32667/nu1.mnc orig_nu.mgz --like orig.mgz --conform
mri_convert.bin ./tmp.mri_nu_correct.mni.32667/nu1.mnc orig_nu.mgz --like orig.mgz --conform
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from ./tmp.mri_nu_correct.mni.32667/nu1.mnc...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
INFO: transform src into the like-volume: orig.mgz
changing data type from float to uchar (noscale = 0)...
MRIchangeType: Building histogram
writing to orig_nu.mgz...
Wed Jul 18 00:11:58 UTC 2018
mri_nu_correct.mni done
talairach_avi --i orig_nu.mgz --xfm transforms/talairach.auto.xfm --atlas 3T18yoSchwartzReactN32_as_orig
talairach_avi log file is transforms/talairach_avi.log...
Started at Wed Jul 18 00:11:58 UTC 2018
Ended at Wed Jul 18 00:12:22 UTC 2018
talairach_avi done
cp transforms/talairach.auto.xfm transforms/talairach.xfm
#--------------------------------------------
#@# Talairach Failure Detection Wed Jul 18 00:12:24 UTC 2018
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
talairach_afd -T 0.005 -xfm transforms/talairach.xfm
ERROR: talairach_afd: Talairach Transform: transforms/talairach.xfm ***FAILED*** (p=0.0614, pval=0.0034 < threshold=0.0050)
Manual Talairach alignment may be necessary, or
include the -notal-check flag to skip this test,
making sure the -notal-check flag follows -all
or -autorecon1 in the command string.
See:
http://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/Talairach
INFO: Trying MINC mritotal to perform Talairach align...
#--------------------------------------------
#@# Talairach Wed Jul 18 00:12:24 UTC 2018
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
mri_nu_correct.mni --no-rescale --i orig.mgz --o orig_nu.mgz --n 1 --proto-iters 1000 --distance 50
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
/opt/freesurfer/bin/mri_nu_correct.mni
--no-rescale --i orig.mgz --o orig_nu.mgz --n 1 --proto-iters 1000 --distance 50
nIters 1
$Id: mri_nu_correct.mni,v 1.27 2016/02/26 16:19:49 mreuter Exp $
Linux node643 3.10.0-693.11.6.el7.x86_64 #1 SMP Thu Jan 4 01:06:37 UTC 2018 x86_64 x86_64 x86_64 GNU/Linux
Wed Jul 18 00:12:24 UTC 2018
Program nu_correct, built from:
Package MNI N3, version 1.12.0, compiled by nicks@terrier (x86_64-unknown-linux-gnu) on 2015-06-19 at 01:25:34
/usr/bin/bc
tmpdir is ./tmp.mri_nu_correct.mni.3455
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
mri_convert orig.mgz ./tmp.mri_nu_correct.mni.3455/nu0.mnc -odt float
mri_convert.bin orig.mgz ./tmp.mri_nu_correct.mni.3455/nu0.mnc -odt float
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from orig.mgz...
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
changing data type from uchar to float (noscale = 0)...
writing to ./tmp.mri_nu_correct.mni.3455/nu0.mnc...
--------------------------------------------------------
Iteration 1 Wed Jul 18 00:12:26 UTC 2018
nu_correct -clobber ./tmp.mri_nu_correct.mni.3455/nu0.mnc ./tmp.mri_nu_correct.mni.3455/nu1.mnc -tmpdir ./tmp.mri_nu_correct.mni.3455/0/ -iterations 1000 -distance 50
[user@node643:/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri/] [2018-07-18 00:12:26] running:
/opt/freesurfer/mni/bin/nu_estimate_np_and_em -parzen -log -sharpen 0.15 0.01 -iterations 1000 -stop 0.001 -shrink 4 -auto_mask -nonotify -b_spline 1.0e-7 -distance 50 -quiet -execute -clobber -nokeeptmp -tmpdir ./tmp.mri_nu_correct.mni.3455/0/ ./tmp.mri_nu_correct.mni.3455/nu0.mnc ./tmp.mri_nu_correct.mni.3455/nu1.imp
Processing:.................................................................Done
Processing:.................................................................Done
Processing:.................................................................Done
Number of iterations: 76
CV of field change: 0.000995587
mri_convert ./tmp.mri_nu_correct.mni.3455/nu1.mnc orig_nu.mgz --like orig.mgz --conform
mri_convert.bin ./tmp.mri_nu_correct.mni.3455/nu1.mnc orig_nu.mgz --like orig.mgz --conform
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from ./tmp.mri_nu_correct.mni.3455/nu1.mnc...
TR=0.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
INFO: transform src into the like-volume: orig.mgz
changing data type from float to uchar (noscale = 0)...
MRIchangeType: Building histogram
writing to orig_nu.mgz...
Wed Jul 18 00:13:56 UTC 2018
mri_nu_correct.mni done
talairach --i orig_nu.mgz --xfm transforms/talairach.auto.xfm
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
/opt/freesurfer/bin/talairach
--i orig_nu.mgz --xfm transforms/talairach.auto.xfm
$Id: talairach,v 1.8 2016/02/16 17:17:20 zkaufman Exp $
Linux node643 3.10.0-693.11.6.el7.x86_64 #1 SMP Thu Jan 4 01:06:37 UTC 2018 x86_64 x86_64 x86_64 GNU/Linux
Wed Jul 18 00:13:56 UTC 2018
tmpdir is transforms/tmp.talairach.4800
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
mri_convert orig_nu.mgz transforms/tmp.talairach.4800/src.mnc
mri_convert.bin orig_nu.mgz transforms/tmp.talairach.4800/src.mnc
$Id: mri_convert.c,v 1.226 2016/02/26 16:15:24 mreuter Exp $
reading from orig_nu.mgz...
TR=1000.00, TE=0.00, TI=0.00, flip angle=0.00
i_ras = (-1, 0, 0)
j_ras = (0, 0, -1)
k_ras = (0, 1, 0)
writing to transforms/tmp.talairach.4800/src.mnc...
--------------------------------------------
mritotal -verbose -debug -clobber -modeldir /opt/freesurfer/mni/bin/../share/mni_autoreg -protocol icbm transforms/tmp.talairach.4800/src.mnc transforms/talairach.auto.xfm
Transforming slices:................................................................................................................................Done
Copying chunks:......................................................................................................................................................Done.
Copying chunks:..........................................................................................................Done.
Transforming slices:......................................................................................................................................................Done
Copying chunks:....................................................................................................................Done.
Copying chunks:....................................................................................................................Done.
Source volume size: 74 by 106 by 106
Source voxel size = 2.000 2.000 2.000
Source min/max real range = -0.000 187.807
Source min/max voxel= -0.000 187.807
Target volume size: 58 by 110 by 86
Target voxel = 2.000 2.000 2.000
Target min/max real range= 0.970 123.710
Target min/max voxel = -0.000 123.710
AFTER init_params()
Transform matrix = 1.0000 0.0000 0.0000 106.0939
0.0000 1.0000 0.0000 -143.6382
0.0000 0.0000 1.0000 -98.6123
Transform center = -105.766 126.047 108.067
Transform rotations = 0.000 0.000 0.000
Transform trans = 106.094 -143.638 -98.612
Transform scale = 1.000 1.000 1.000
Transform shear = 0.000 0.000 0.000
Target volume is smallest
Lattice step size = 4.000 4.000 4.000
Lattice start = -85.095 -125.510 -47.250
Lattice count = 43 55 29
Source volume size: 84 by 116 by 116
Source voxel size = 2.000 2.000 2.000
Source min/max real range = -0.000 228.534
Source min/max voxel= -0.000 228.534
Target volume size: 68 by 110 by 86
Target voxel = 2.000 2.000 2.000
Target min/max real range= -0.000 144.070
Target min/max voxel = -0.000 144.070
AFTER init_params()
Transform matrix = 0.7569 -0.1210 0.2308 73.0487
-0.0273 0.6683 0.4399 -124.7935
-0.2592 -0.4238 0.6277 -39.4539
Transform center = -105.766 126.047 108.067
Transform rotations = -0.594 0.330 -0.036
Transform trans = 108.453 -116.173 -105.694
Transform scale = 0.801 0.801 0.801
Transform shear = 0.000 0.000 0.000
Target volume is smallest
Lattice step size = 4.000 4.000 4.000
Lattice start = -85.095 -125.510 -57.250
Lattice count = 43 55 34
Source volume size: 84 by 116 by 116
Source voxel size = 2.000 2.000 2.000
Source min/max real range = 0.018 22.809
Source min/max voxel= 0.018 22.809
Target volume size: 68 by 110 by 86
Target voxel = 2.000 2.000 2.000
Target min/max real range= 0.037 11.652
Target min/max voxel = 0.037 11.652
AFTER init_params()
Transform matrix = 0.8808 -0.0365 -0.0515 102.0183
0.0573 0.7643 0.4387 -135.3286
0.0264 -0.4409 0.7647 -22.4860
Transform center = -105.766 126.047 108.067
Transform rotations = -0.523 -0.030 0.065
Transform trans = 104.455 -123.700 -106.280
Transform scale = 0.883 0.883 0.883
Transform shear = 0.000 -0.000 -0.000
Target volume is smallest
Lattice step size = 4.000 4.000 4.000
Lattice start = -65.095 -105.510 -57.250
Lattice count = 35 46 34
Source volume size: 84 by 116 by 116
Source voxel size = 2.000 2.000 2.000
Source min/max real range = 0.018 22.809
Source min/max voxel= 0.018 22.809
Target volume size: 68 by 110 by 86
Target voxel = 2.000 2.000 2.000
Target min/max real range= 0.037 11.652
Target min/max voxel = 0.037 11.652
AFTER init_params()
Transform matrix = 0.6988 -0.0762 -0.0240 81.7916
0.0782 0.6093 0.3425 -94.6126
-0.0163 -0.3430 0.6139 -25.0363
Transform center = -105.766 126.047 108.067
Transform rotations = -0.510 0.023 0.111
Transform trans = 101.446 -115.115 -108.267
Transform scale = 0.703 0.703 0.703
Transform shear = -0.000 0.000 0.000
Target volume is smallest
Lattice step size = 4.000 4.000 4.000
Lattice start = -65.095 -105.510 -57.250
Lattice count = 35 46 34
Source volume size: 84 by 116 by 116
Source voxel size = 2.000 2.000 2.000
Source min/max real range = 0.018 22.809
Source min/max voxel= 0.018 22.809
Target volume size: 68 by 110 by 86
Target voxel = 2.000 2.000 2.000
Target min/max real range= 0.037 11.652
Target min/max voxel = 0.037 11.652
AFTER init_params()
Transform matrix = 0.6734 -0.1473 -0.0791 91.8724
0.2231 0.7243 0.5505 -124.0833
-0.0299 -0.4871 0.6531 -28.8442
Transform center = -105.766 126.047 108.067
Transform rotations = -0.641 0.037 0.241
Transform trans = 99.303 -122.938 -124.582
Transform scale = 0.694 0.937 0.815
Transform shear = 0.000 0.000 0.000
Target volume is smallest
Lattice step size = 4.000 4.000 4.000
Lattice start = -65.095 -105.510 -57.250
Lattice count = 35 46 34
Source volume size: 84 by 116 by 116
Source voxel size = 2.000 2.000 2.000
Source min/max real range = 0.018 22.809
Source min/max voxel= 0.018 22.809
Target volume size: 68 by 110 by 86
Target voxel = 2.000 2.000 2.000
Target min/max real range= 0.037 11.652
Target min/max voxel = 0.037 11.652
AFTER init_params()
Transform matrix = 0.6697 -0.1395 -0.1210 94.6509
0.2435 0.6481 0.6006 -116.7125
-0.0078 -0.6267 0.6795 -6.0976
Transform center = -105.766 126.047 108.067
Transform rotations = -0.745 0.008 0.269
Transform trans = 98.919 -121.927 -118.908
Transform scale = 0.695 0.917 0.924
Transform shear = -0.000 0.000 -0.000
Target volume is smallest
Lattice step size = 4.000 4.000 4.000
Lattice start = -65.095 -105.510 -57.250
Lattice count = 35 46 34
Wed Jul 18 00:14:06 UTC 2018
talairach done
cp transforms/talairach.auto.xfm transforms/talairach.xfm
#--------------------------------------------
#@# Talairach Failure Detection Wed Jul 18 00:14:08 UTC 2018
/home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/mri
talairach_afd -T 0.005 -xfm transforms/talairach.xfm
ERROR: talairach_afd: Talairach Transform: transforms/talairach.xfm ***FAILED*** (p=0.0455, pval=0.0034 < threshold=0.0050)
Manual Talairach alignment may be necessary, or
include the -notal-check flag to skip this test,
making sure the -notal-check flag follows -all
or -autorecon1 in the command string.
See:
http://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/Talairach
ERROR: Talairach failed!
Linux node643 3.10.0-693.11.6.el7.x86_64 #1 SMP Thu Jan 4 01:06:37 UTC 2018 x86_64 x86_64 x86_64 GNU/Linux
recon-all -s sub-0180 exited with ERRORS at Wed Jul 18 00:14:08 UTC 2018
For more details, see the log file /home/user/study/PHASE2/input_data/OAS1_bids/derivatives/fmriprep_1.1.1/freesurfer/sub-0180/scripts/recon-all.log
To report a problem, see http://surfer.nmr.mgh.harvard.edu/fswiki/BugReporting
Standard error:
Return code: 1