Dear fmriprep experts,
Thanks for your help in advance!
I have some single-band fMRI data and a corresponding field map (in Hz) generated directly from a GE scanner, below please see the folder structure.
└── ses-02
├── anat
│ ├── sub-IRIS009_ses-02_T1w.json
│ └── sub-IRIS009_ses-02_T1w.nii.gz
├── fmap
│ ├── sub-IRIS009_ses-02_run-00_fieldmap.json
│ ├── sub-IRIS009_ses-02_run-00_fieldmap.nii.gz
│ └── sub-IRIS009_ses-02_run-00_magnitude.nii.gz
└── func
├── sub-IRIS009_ses-02_task-conscious_acq-sb_dir-pe1_run-00_bold.json
├── sub-IRIS009_ses-02_task-conscious_acq-sb_dir-pe1_run-00_bold.nii.gz
├── sub-IRIS009_ses-02_task-rs_acq-sb_dir-pe1_run-00_bold.json
└── sub-IRIS009_ses-02_task-rs_acq-sb_dir-pe1_run-00_bold.nii.gz
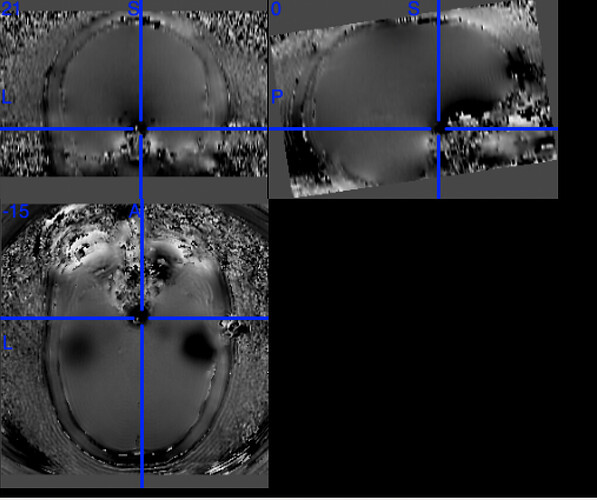
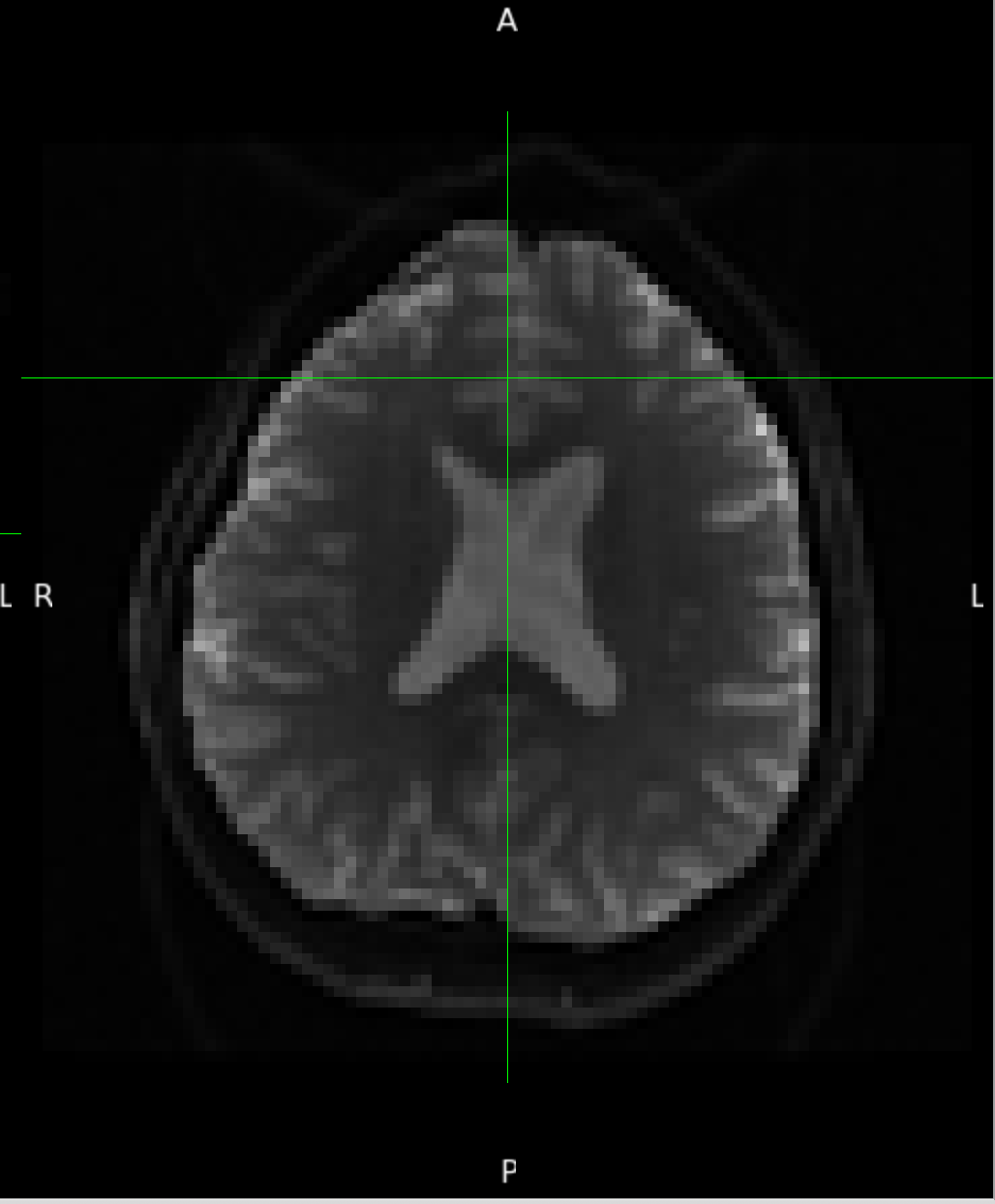
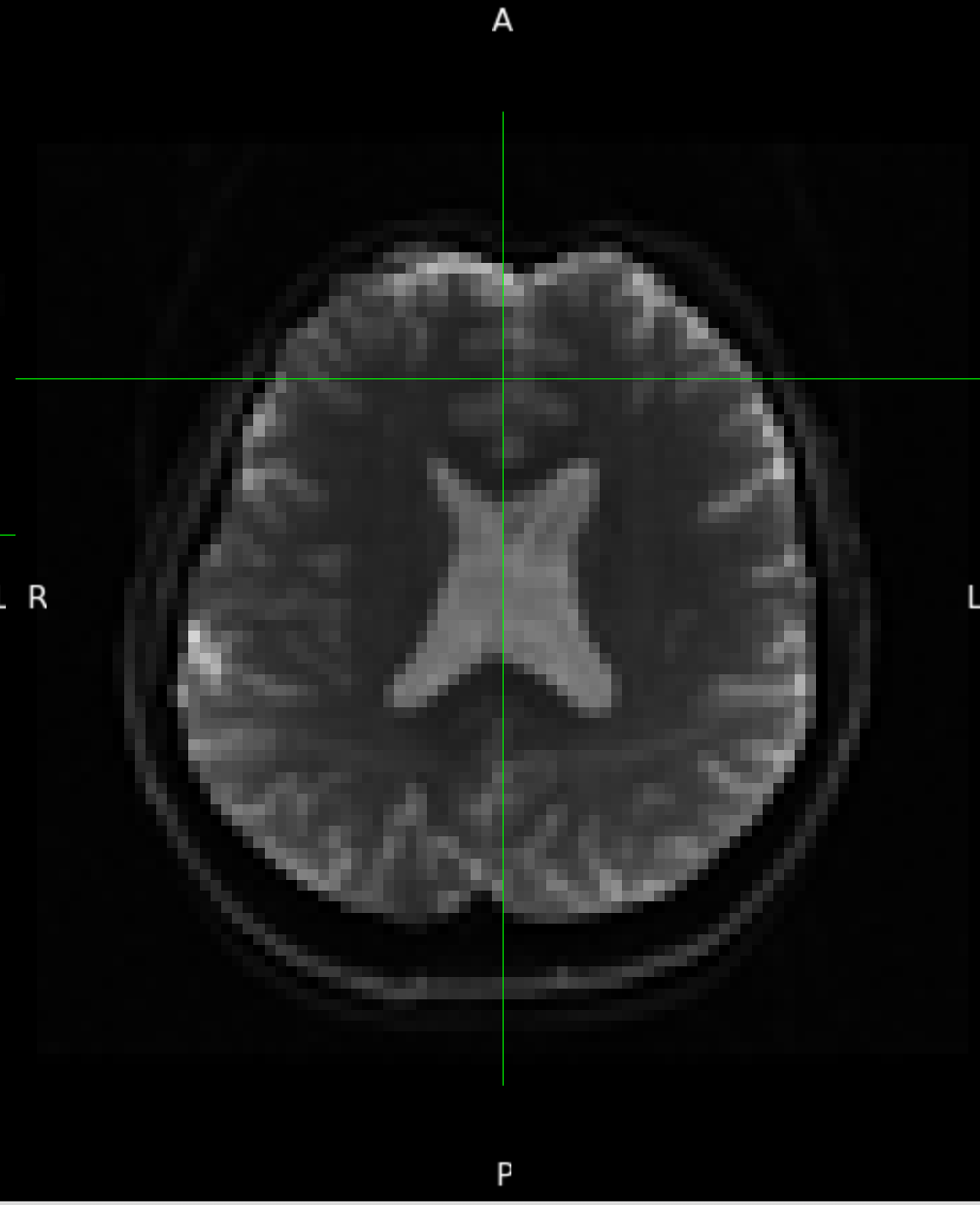
One example of the scanner-generated field map.
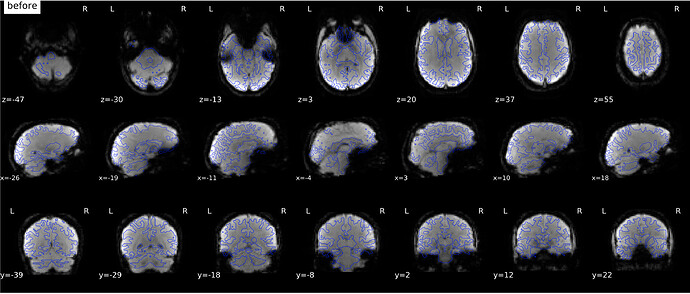
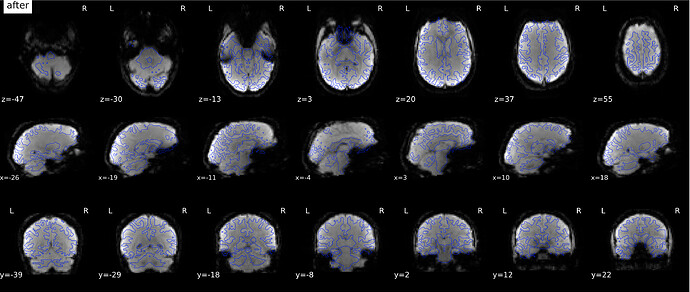
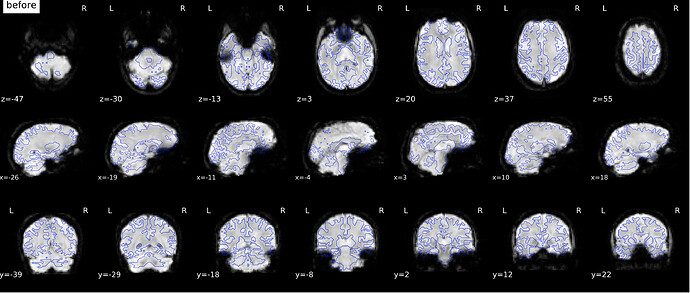
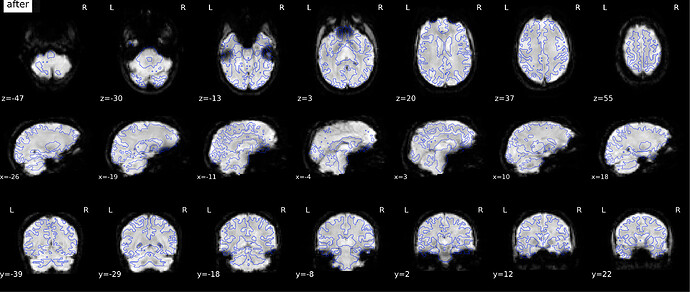
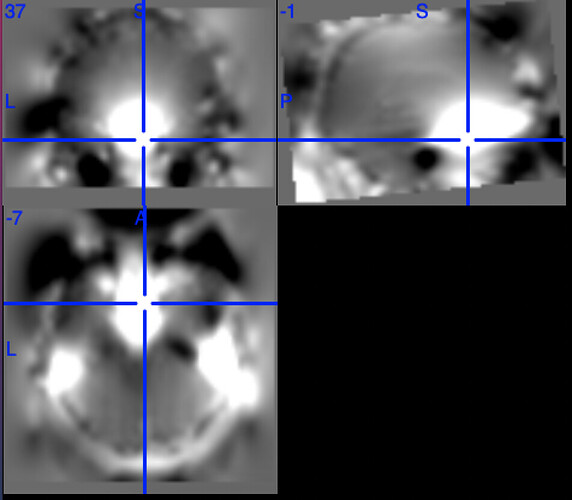
According to the summary report, fMRIprep used FMB (fieldmap-based) - directly measured B0 map in the SDC. Somehow the images after susceptibility distortion correction (SDC) in fMRIPrep 20.2.3 were even worse - more distortion, especially in the medial frontal area and the brainstem (see below). Sorry, it’s less obvious than a gif.
I checked the phase encoding direction of the fMRI data and it was correctly written in the .json sidecar - here it’s AP, our Nifti files (the functional data and the field map) are in LAS orientation, so PhaseEncodingDirection = j-.
If I flip the phase encoding direction to PA, so PhaseEncodingDirection = j, the correction seems correct, however, based on the imaging protocol and the distortion, the phase encoding direction should be AP instead of PA.
I wonder,
(1) if the field map should be somehow multiplied by -1 (flipped its sign) to be entered into fmriprep - I tried to do so and the corrected images looks the same as flipping the PE direction.
(2) if fMRIprep reorients functional images during/before SDC correction, but does not change the PhaseEncodingDirection.
Any suggestion will be appreciated!