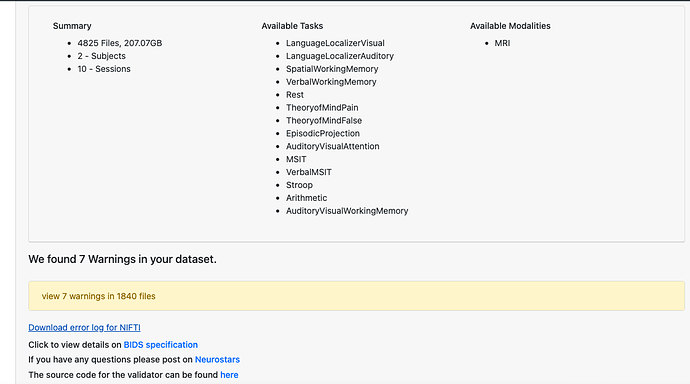
Summary of what happened:
I am receiving the below error using fmriprep version 21.0.2. I have used this version successfully before, but this data is from a new scanner which we haven’t preprocessed before, so it’s probably related to that.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv -B /projects/b1081:/projects/b1081 \
-B ${WORK_DIR}:/quest_scratch \
/projects/b1081/singularity_images/fmriprep-21.0.2.simg \
${BIDS_DIR} \
${BIDS_DIR}/${DERIVS_DIR} \
participant --participant-label ${subject} \
-w /quest_scratch --omp-nthreads 8 --nthreads 16 \
--fs-license-file /projects/b1081/singularity_images/freesurfer_license.txt \
--fs-subjects-dir ${BIDS_DIR}/${DERIVS_DIR}/sourcedata/freesurfer \
--output-spaces MNI152NLin6Asym:res-2 \
--ignore slicetiming --fd-spike-threshold 0.2 --me-output-echos --resource-monitor
Version: 21.0.2
Environment (Docker, Singularity, custom installation): singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
Yes, validated w BIDS validator.Relevant log outputs (up to 20 lines):
Node: fmriprep_wf.single_subject_HUBS02_wf.fmap_preproc_wf.wf_auto_00003.bs_filter
Working directory: /quest_scratch/fmriprep_wf/single_subject_HUBS02_wf/fmap_preproc_wf/wf_auto_00003/bs_filter
Node inputs:
bs_spacing = [(100.0, 100.0, 40.0), (16.0, 16.0, 10.0)]
extrapolate = True
in_data = <undefined>
in_mask = <undefined>
recenter = mode
ridge_alpha = 0.01
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 516, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 635, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 741, in _run_command
result = self._interface.run(cwd=outdir)
File "/opt/conda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 428, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.8/site-packages/sdcflows/interfaces/bspline.py", line 141, in _run_interface
data -= mode(data[mask], axis=None)[0][0]
IndexError: boolean index did not match indexed array along dimension 2; dimension is 4 but corresponding boolean dimension is 54