Summary of what happened:
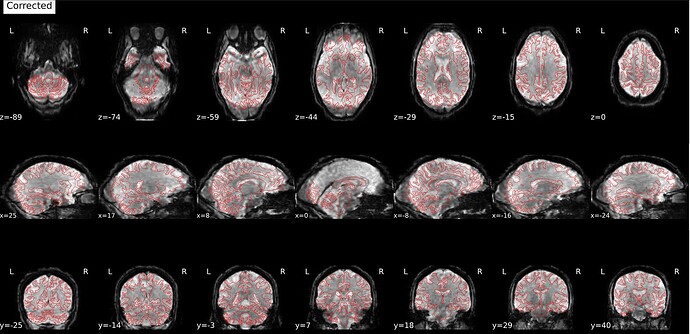
Hi everyone! I’m currently trying to use fMRIPrep to preprocess a dataset from openneuro (ds006072). fMRIPrep is running smoothly in general, but there seems to be a problem with SDC, which leads to a distorted and elongated corrected image. This is one example, but the data for all my sessions is similarly distorted:
The JSON sidecards for the fmap data were missing the B0FieldIdentifier or IntendedFor fields. I checked other threads here for multi-echo data, so I only linked to the first echo of their respectives files. I also checked that the dataset_description.json has set “DatasetType”: “raw”. Here is how I added the IntendedFor fields once for AP:
"IntendedFor": [
"ses-1/func/sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-1_part-mag_bold.nii",
"ses-1/func/sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-1_part-mag_bold.nii"
]
and once for PA:
"IntendedFor": [
"ses-1/func/sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-1_part-mag_bold.nii",
"ses-1/func/sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-1_part-mag_bold.nii"
]
and here is the file tree:
sub-P3
ses-1
anat
sub-P3_ses-1_acq-t1mpr0p9mm_rec-NDNORM_run-1_T1w.json
sub-P3_ses-1_acq-t1mpr0p9mm_rec-NDNORM_run-1_T1w.nii.gz
sub-P3_ses-1_acq-t1mpr0p9mm_rec-NDNORM_run-2_T1w.json
sub-P3_ses-1_acq-t1mpr0p9mm_rec-NDNORM_run-2_T1w.nii.gz
sub-P3_ses-1_acq-t2spc0p9mmiso_rec-NDNORM_run-1_T2w.json
sub-P3_ses-1_acq-t2spc0p9mmiso_rec-NDNORM_run-1_T2w.nii.gz
sub-P3_ses-1_acq-t2spc0p9mmiso_rec-NDNORM_run-2_T2w.json
sub-P3_ses-1_acq-t2spc0p9mmiso_rec-NDNORM_run-2_T2w.nii.gz
dwi
fmap
sub-P3_ses-1_acq-SpinEchoFieldMapAP_dir-AP_run-1_epi.json
sub-P3_ses-1_acq-SpinEchoFieldMapAP_dir-AP_run-1_epi.nii
sub-P3_ses-1_acq-SpinEchoFieldMapAP_dir-AP_run-2_epi.json
sub-P3_ses-1_acq-SpinEchoFieldMapAP_dir-AP_run-2_epi.nii
sub-P3_ses-1_acq-SpinEchoFieldMapPA_dir-PA_run-1_epi.json
sub-P3_ses-1_acq-SpinEchoFieldMapPA_dir-PA_run-1_epi.nii
sub-P3_ses-1_acq-SpinEchoFieldMapPA_dir-PA_run-2_epi.json
sub-P3_ses-1_acq-SpinEchoFieldMapPA_dir-PA_run-2_epi.nii
func
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-1_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-1_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-2_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-2_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-3_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-3_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-4_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-4_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-5_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-1_echo-5_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-1_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-1_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-2_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-2_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-3_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-3_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-4_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-4_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-5_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST1_dir-AP_run-2_echo-5_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-1_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-1_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-2_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-2_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-3_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-3_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-4_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-4_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-5_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-1_echo-5_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-1_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-1_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-2_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-2_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-3_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-3_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-4_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-4_part-mag_bold.nii
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-5_part-mag_bold.json
sub-P3_ses-1_task-BOLDREST2_dir-AP_run-2_echo-5_part-mag_bold.nii
Command used (and if a helper script was used, a link to the helper script or the command generated):
BIDS_DIR="/mnt/d/Tim_MA/rawdata"
OUT_DIR="/mnt/d/Tim_MA/derivatives"
FILTER_FILE="/mnt/d/Tim_MA/bids_filters.json"
SUBJECT="$1" # pass subject as argument
# Create a unique work directory for each subject
WORK_DIR="/mnt/d/Tim_MA/work/${SUBJECT}"
mkdir -p "${WORK_DIR}"
DOCKER_IMAGE="nipreps/fmriprep:latest"
NCPUS=8
MEM_GB=32
MEM_MB=$((MEM_GB*1000-500))
export TEMPLATEFLOW_HOME=$HOME/.cache/templateflow
echo "Processing ${SUBJECT}..."
docker run -ti --rm \
-v ${BIDS_DIR}:/data:ro \
-v ${OUT_DIR}:/out \
-v ${WORK_DIR}:/scratch \
-v ${FILTER_FILE}:/bids_filters.json:ro \
${DOCKER_IMAGE} /data /out participant \
--participant-label ${SUBJECT} \
--fs-license-file /out/license.txt \
--bids-filter-file /bids_filters.json \
--work-dir /scratch \
--output-spaces MNI152NLin2009cAsym \
--nprocs ${NCPUS} \
--mem ${MEM_MB} \
--skip_bids_validation \
--stop-on-first-crash
Version:
25.2.3
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
On openneuro.org it says that the data is BIDS valid, but missing some optional flags, like B0FieldIdentifier.
Relevant log outputs (up to 20 lines):
fMRIPrep is running without any error messages.
Screenshots / relevant information:
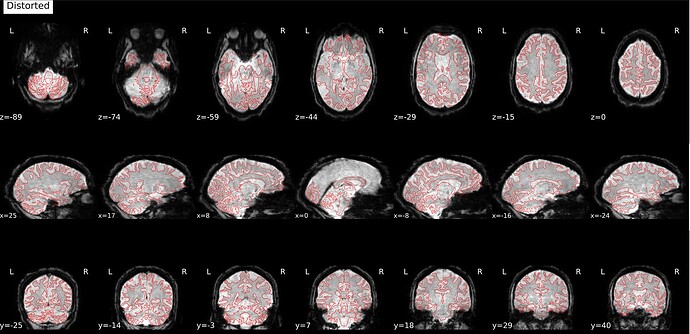
This is what the distorted version looks like, before SDC:
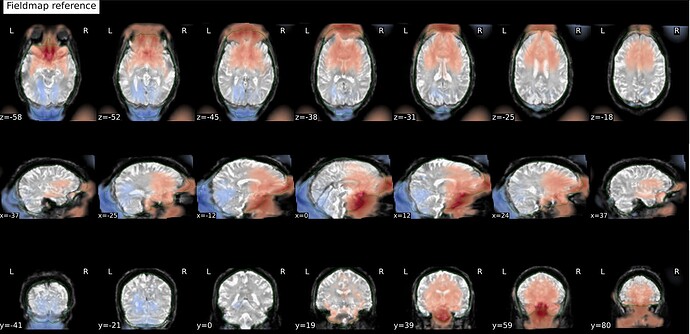
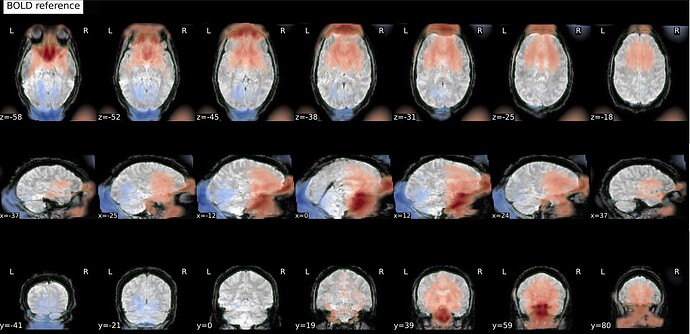
And these are the fmapCoreg_bold images:
I also only want the resting-state scans preprocessed to save some time, so I used this bids_filter:
{
"bold": {
"datatype": "func",
"suffix": "bold",
"task": [
"BOLDREST1",
"BOLDREST2",
"BOLDREST3",
"BOLDREST4",
"BOLDREST5",
"BOLDREST6"
]
}
}
Thanks for the help!