Summary of what happened:
Hello everyone,
I am new to fMRIPrep and I want to use it to preprocess my fMRI data. My data is in BIDS format (validated using BIDS Validator). I am using a 3D multi-echo (3 echoes) sequence with 2 sessions and 4 runs each. Before every functional run (AP direction), I run a short fmap in PA direction. My multi-echo sequence produces 3 echo NIfTI files per sequence. So for each functional run with a task, I have:
func
sub-HM70_ses-01_task-pi_run-01_echo-01_bold.nii.gz
sub-HM70_ses-01_task-pi_run-01_echo-02_bold.nii.gz
sub-HM70_ses-01_task-pi_run-01_echo-03_bold.nii.gz
fmap
sub-HM70_ses-01_acq-echo01_dir-PA_run-01_epi.nii.gz
sub-HM70_ses-01_acq-echo02_dir-PA_run-01_epi.nii.gz
sub-HM70_ses-01_acq-echo03_dir-PA_run-01_epi.nii.gz
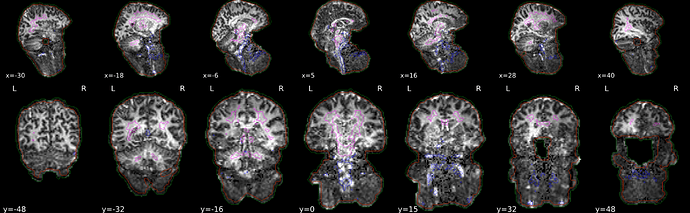
The below command was running fine without erros but produced weird looking output under the functional tab for “Brain mask and (anatomical/temporal) CompCor ROIs” (see screenshot).
As far as I could interpret this, brain was detected at the neck area… Is this normal or can I ignore this?
I thought that I could improve the output by including my fieldmaps. So far I did not have the IntendedFor key in the json file of my fmaps, but then I wanted to add this. This led to some errors (topup crash, extracted from log file), thus I did not have an output at all:
According to another conversation (Multi echo data field maps for fmriprep - #2 by Steven), I referenced my 3 func echos to echo01 of my fmap (see option a below).
My question now concerns the referencing of fmap and func, because I think there are several options:
option a: in json fmap echo01 I reference all 3 echoes of func → 1 to 3 mapping:
"IntendedFor": [
"ses-01/func/sub-HM70_ses-01_task-pi_run-01_echo-01_bold.nii.gz",
"ses-01/func/sub-HM70_ses-01_task-pi_run-01_echo-02_bold.nii.gz",
"ses-01/func/sub-HM70_ses-01_task-pi_run-01_echo-03_bold.nii.gz"
]
→ I tried this and it always produced an topup crash
option b: in json fmap echo01 I reference echo01 of func (and in json fmap echo02 I reference func echo02, in json fmap echo03 I reference func echo03) → 1:1 mapping:
"IntendedFor": [
"ses-01/func/sub-HM70_ses-01_task-pi_run-01_echo-01_bold.nii.gz",
]
What option should I use and why?
Is there anything else I could improve or try?
I am grateful for any help! ![]()
Command used (and if a helper script was used, a link to the helper script or the command generated):
I sucessfully ran a “basic” fmriprep command on our HPC cluster:
apptainer run \
--env APPTAINERENV_TEMPLATEFLOW_HOME=/tmp/templateflow \
-B /lustre/myuser/rawdata/BIDS:/bids \
-B /lustre/myuser/preprocessing/fmriprep:/out \
/lustre/myuser/preprocessing/resources/fmriprep_latest.sif \
--participant-label HM70 \
--output-spaces T1w OASIS30ANTs MNI152NLin2009cAsym \
--nthreads 16 \
--omp-nthreads 8 \
--mem-mb 60000 \
--fs-license-file /tmp/license.txt \
/bids /out participant
Version:
25.2.3, and also tried 25.2.4
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
For BIDS validation I used: https://bids-standard.github.io/bids-validator/
This showed no errors, but the following warnings:
warning: README_FILE_MISSING
warning: TOO_FEW_AUTHORS
warning: JSON_KEY_RECOMMENDED (HEDVersion)
warning: JSON_KEY_RECOMMENDED (GeneratedBy)
warning: JSON_KEY_RECOMMENDED (SourceDatasets)
warning: SIDECAR_KEY_RECOMMENDED (MatrixCoilMode)
warning: SIDECAR_KEY_RECOMMENDED (PulseSequenceType)
warning: SIDECAR_KEY_RECOMMENDED (SpoilingType)
warning: SIDECAR_KEY_RECOMMENDED (PartialFourierDirection)
warning: SIDECAR_KEY_RECOMMENDED (Instructions)
warning: SIDECAR_KEY_RECOMMENDED (CogAtlasID)
warning: SIDECAR_KEY_RECOMMENDED (CogPOID)
warning: EVENTS_TSV_MISSING
Relevant log outputs (up to 20 lines):
260204-18:26:36,398 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00000.topup
260204-18:26:36,401 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00003.topup
260204-18:26:36,404 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00006.topup
260204-18:26:36,406 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00009.topup
260204-18:26:36,408 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00012.topup
260204-18:26:36,410 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00015.topup
260204-18:26:36,412 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00018.topup
260204-18:26:36,415 nipype.workflow ERROR: could not run node: fmriprep_25_2_wf.sub_HM70_ses_01-02_wf.fmap_preproc_wf.wf_auto_00021.topup
260204-18:26:36,456 nipype.workflow CRITICAL: fMRIPrep failed: 8 raised. Re-raising first.