Summary of what happened:
Has anyone encountered this kind of issue when processing T1w images with FreeSurfer? I ran recon-all using Freesurfer 7.3.2 and fed the images to fMRIPrep 25.1.4. The images look identical to those generated when recon-all is run directly within fMRIPrep.
Command used (and if a helper script was used, a link to the helper script or the command generated):
PASTE CODE HERE
Version:
25.1.4
Environment (Docker, Singularity / Apptainer, custom installation):
PUT ENVIRONMENT HERE
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE
Screenshots / relevant information:
Spatial normalization
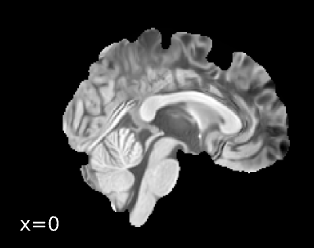
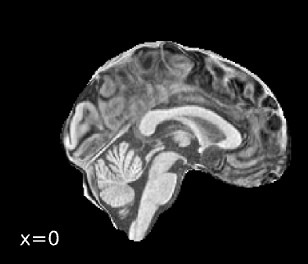
And below is the boilerplate text from the FMRIPrep summary report:
Anatomical data preprocessing
: A total of 1 T1-weighted (T1w) images were found within the input
BIDS dataset. The T1w image was corrected for intensity
non-uniformity (INU) withN4BiasFieldCorrection[@n4], distributed with ANTs 2.6.2
[@ants, RRID:SCR_004757], and used as T1w-reference throughout the workflow.
The T1w-reference was then skull-stripped with a Nipype implementation of
theantsBrainExtraction.shworkflow (from ANTs), using MNI152NLin2009cAsym
as target template.
Brain tissue segmentation of cerebrospinal fluid (CSF),
white-matter (WM) and gray-matter (GM) was performed on
the brain-extracted T1w usingfast[FSL (version unknown), RRID:SCR_002823, @fsl_fast].
Brain surfaces were reconstructed usingrecon-all[FreeSurfer 7.3.2,
RRID:SCR_001847, @fs_reconall], and the brain mask estimated
previously was refined with a custom variation of the method to reconcile
ANTs-derived and FreeSurfer-derived segmentations of the cortical
gray-matter of Mindboggle [RRID:SCR_002438, @mindboggle].
A T2-weighted image was used to improve pial surface refinement.
Brain surfaces were reconstructed usingrecon-all[FreeSurfer 7.3.2,
RRID:SCR_001847, @fs_reconall], and the brain mask estimated
previously was refined with a custom variation of the method to reconcile
ANTs-derived and FreeSurfer-derived segmentations of the cortical
gray-matter of Mindboggle [RRID:SCR_002438, @mindboggle].
Volume-based spatial normalization to two standard spaces (MNI152NLin2009cAsym, MNI152NLin6Asym) was performed through
nonlinear registration withantsRegistration(ANTs 2.6.2),
using brain-extracted versions of both T1w reference and the T1w template.
The following templates were were selected for spatial normalization
and accessed with TemplateFlow [24.2.2, @templateflow]:
ICBM 152 Nonlinear Asymmetrical template version 2009c [@mni152nlin2009casym, RRID:SCR_008796; TemplateFlow ID: MNI152NLin2009cAsym], FSL’s MNI ICBM 152 non-linear 6th Generation Asymmetric Average Brain Stereotaxic Registration Model [@mni152nlin6asym, RRID:SCR_002823; TemplateFlow ID: MNI152NLin6Asym].
Grayordinate “dscalar” files containing 91k samples were
resampled onto fsLR using the Connectome Workbench [@hcppipelines].

