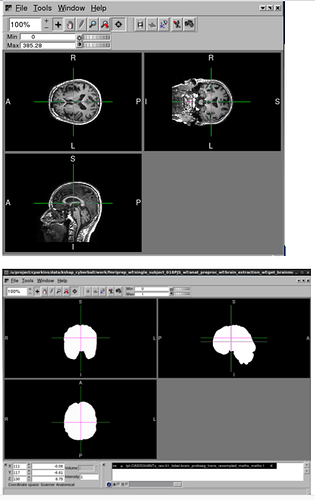
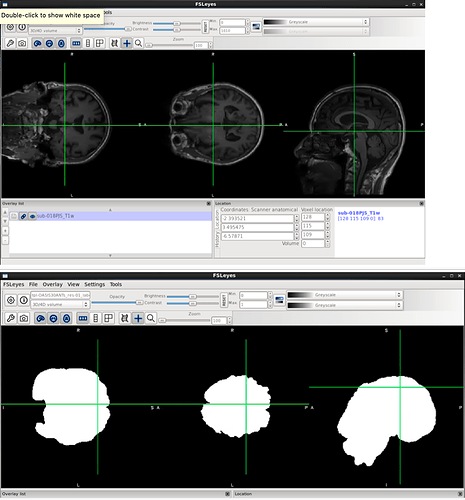
Hello, I have a dataset that I’ve run through fMRIPrep where about 20 of the subjects (out of 126) crashed with similar errors.
I have tried to re-run the subjects with increased memory allocation, but it continues to crash with the same error. Any help would be greatly appreciated. Thank you!
The error at the end of the HTML output file of one of the subjects:
Node Name: fmriprep_wf.single_subject_018PJS_wf.anat_preproc_wf.brain_extraction_wf.atropos_wf.01_atropos
The error in the log:
Node: fmriprep_wf.single_subject_018PJS_wf.anat_preproc_wf.brain_extraction_wf.atropos_wf.01_atropos
Working directory: /u/project/cparkins/data/kshap_cyberball/work/fmriprep_wf/single_subject_018PJS_wf/anat_preproc_wf/brain_extraction_wf/atropos_wf/01_atropos
Node inputs:
args =
convergence_threshold = 0.0
dimension = 3
environ = {‘NSLOTS’: ‘8’}
icm_use_synchronous_update =
initialization = KMeans
intensity_images = [’/u/project/cparkins/data/kshap_cyberball/work/fmriprep_wf/single_subject_018PJS_wf/anat_preproc_wf/brain_extraction_wf/inu_n4/mapflow/inu_n40/sub-018PJS_T1w_ras_valid_maths_corrected.nii.gz’]
likelihood_model = Gaussian
mask_image = /u/project/cparkins/data/kshap_cyberball/work/fmriprep_wf/single_subject_018PJS_wf/anat_preproc_wf/brain_extraction_wf/get_brainmask/tpl-OASIS30ANTs_res-01_label-brain_probseg_trans_resampled_maths_maths.nii.gz
maximum_number_of_icm_terations =
mrf_radius = [1, 1, 1]
mrf_smoothing_factor = 0.1
n_iterations = 3
num_threads = 8
number_of_tissue_classes = 3
out_classified_image_name =
output_posteriors_name_template = POSTERIOR%02d.nii.gz
posterior_formulation =
prior_probability_images =
prior_probability_threshold =
prior_weighting =
save_posteriors =
use_mixture_model_proportions =
use_random_seed = True
Traceback (most recent call last):
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/pipeline/plugins/multiproc.py”, line 69, in run_node
result[‘result’] = node.run(updatehash=updatehash)
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py”, line 472, in run
result = self._run_interface(execute=True)
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py”, line 563, in _run_interface
return self._run_command(execute)
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/pipeline/engine/nodes.py”, line 643, in _run_command
result = self._interface.run(cwd=outdir)
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/interfaces/base/core.py”, line 375, in run
runtime = self._run_interface(runtime)
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/interfaces/ants/segmentation.py”, line 163, in _run_interface
runtime = super(Atropos, self)._run_interface(runtime)
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/interfaces/base/core.py”, line 758, in _run_interface
self.raise_exception(runtime)
File “/u/project/CCN/apps/fmriprep/1.4.0_py3.7.2/lib/python3.7/site-packages/nipype/interfaces/base/core.py”, line 695, in raise_exception
).format(**runtime.dictcopy()))
RuntimeError: Command:
Atropos --image-dimensionality 3 --initialization KMeans[3] --intensity-image /u/project/cparkins/data/kshap_cyberball/work/fmriprep_wf/single_subject_018PJS_wf/anat_preproc_wf/brain_extraction_wf/inu_n4/mapflow/_inu_n40/sub-018PJS_T1w_ras_valid_maths_corrected.nii.gz --likelihood-model Gaussian --mask-image /u/project/cparkins/data/kshap_cyberball/work/fmriprep_wf/single_subject_018PJS_wf/anat_preproc_wf/brain_extraction_wf/get_brainmask/tpl-OASIS30ANTs_res-01_label-brain_probseg_trans_resampled_maths_maths.nii.gz --mrf [0.1,1x1x1] --convergence [3,0] --output [sub-018PJS_T1w_ras_valid_maths_corrected_labeled.nii.gz] --use-random-seed 1
Standard output:
Standard error:
Return code: 1