Summary of what happened
Dear fMRIPrep,
I’m trying to run a fMRIPrep to preprocess subject data, using this command:
The data_description.json file is:
{"Name":"MS Fatigue study","BIDSVersion":"1.8.0","License":"PDDL","Authors":["Benelli A.","Cortese R.","Tatti E.","Massucco E.","Luchetti L.","Battaglini M.","Cudeiro J.","de Mauro A.","Plantone D.","Pasqualetti P.","Righi D.","Neri F.","Stromillo M.L.","Cinti A.","Giannotta A.","Lomi F.","Scoccia A.","Lai G.","De Stefano N.","Ulivelli M.","Rossi S."],"HowToAcknowledge":"When using this data, please cite Benelli et al.","Funding":"to be defined","ReferencesAndLinks":["to be defined"]}
the task-rest_bold.json file is:
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 127.738237,
"Manufacturer": "Philips",
"ManufacturersModelName": "Achieva dStream",
"InstitutionName": "Ospedale Pediatrico Meyer",
"InstitutionalDepartmentName": "Risonanza Magnetica",
"InstitutionAddress": "Viale Pieraccini 24, Firenze",
"DeviceSerialNumber": "17260",
"StationName": "PHILIPS-P9BMSKQ",
"BodyPart": "BRAIN",
"PatientPosition": "HFS",
"ProcedureStepDescription": "RM ENCEFALO",
"SoftwareVersions": "5.3.1/5.3.1.4",
"MRAcquisitionType": "2D",
"StudyDescription": "RM ENCEFALO",
"SeriesDescription": "FE_EPI SENSE",
"ProtocolName": "FE_EPI SENSE",
"ScanningSequence": "GR",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"PulseSequenceName": "EPI",
"ImageType": ["ORIGINAL", "PRIMARY", "T2", "NONE", "MAGNITUDE"],
"SeriesNumber": 701,
"AcquisitionTime": "15:30:53.300000",
"AcquisitionNumber": 7,
"PhilipsRescaleSlope": 0.951893,
"PhilipsRescaleIntercept": 0,
"PhilipsScaleSlope": 0.0288861,
"UsePhilipsFloatNotDisplayScaling": 1,
"SliceThickness": 4,
"SpacingBetweenSlices": 4,
"TablePosition": [
0,
0,
0.3 ],
"EchoTime": 0.035,
"RepetitionTime": 3,
"MTState": false,
"FlipAngle": 90,
"CoilString": "MULTI COIL",
"PercentPhaseFOV": 100,
"PercentSampling": 98.4375,
"EchoTrainLength": 71,
"PhaseEncodingSteps": 126,
"FrequencyEncodingSteps": 128,
"PhaseEncodingStepsOutOfPlane": 1,
"AcquisitionMatrixPE": 126,
"ReconMatrixPE": 128,
"ParallelReductionFactorInPlane": 1.8,
"ParallelAcquisitionTechnique": "SENSE",
"WaterFatShift": 22.1019,
"EstimatedEffectiveEchoSpacing": 0.00039514,
"EstimatedTotalReadoutTime": 0.0501828,
"AcquisitionDuration": 611.938,
"PixelBandwidth": 2330.35,
"PhaseEncodingAxis": "j",
"ImageOrientationPatientDICOM": [
0.998752,
0.0473536,
0.0158947,
-0.0470147,
0.998672,
-0.0210601 ],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"BidsGuess": ["func","_acq-SKGRFEEPIp18_run-701_bold"],
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20241208"
}
Within the study folder I’ve these subfolders: derivatives (empty), work (empty), sub-01 (with both anat and func subsubfolders); I’ve license.txt also.
Inside both anat and fund subsubfolders there the .gz file and the descriptive .json file, following the correct names for BIDS format.
I don’t know how to fix it.
Is there anyone who can help me woth that?
Best regards,
Alberto
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep \
/Volumes/BENO_ALBE/prova/MS_FATIGUE_BIDS_format \
/Volumes/BENO_ALBE/prova/MS_FATIGUE_BIDS_format/derivatives participant \
-w /Volumes/BENO_ALBE/prova/MS_FATIGUE_BIDS_format/work/
Version:
Environment (Docker, Singularity / Apptainer, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
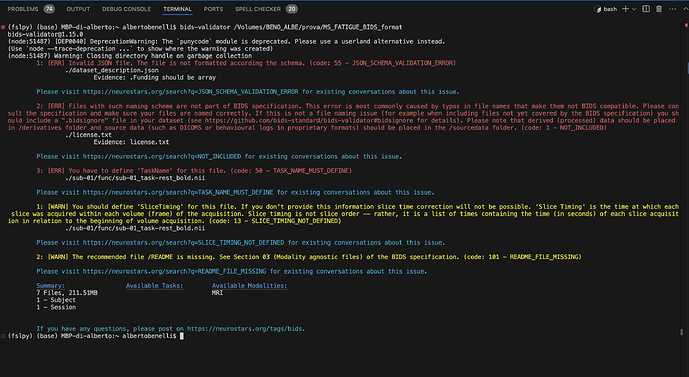
Running bids-validator I get this other error (attached)
Relevant log outputs (up to 20 lines):
When I run it from terminal I get this error:
s/BENO_ALBE/prova/MS_FATIGUE_BIDS_format/work/
241211-16:15:44,887 cli WARNING:
Telemetry disabled because sentry_sdk is not installed.
Traceback (most recent call last):
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/bids/layout/index.py", line 331, in load_json
return json.load(handle)
File "/opt/anaconda3/envs/fslpy/lib/python3.10/json/__init__.py", line 293, in load
return loads(fp.read(),
File "/opt/anaconda3/envs/fslpy/lib/python3.10/codecs.py", line 322, in decode
(result, consumed) = self._buffer_decode(data, self.errors, final)
UnicodeDecodeError: 'utf-8' codec can't decode byte 0xb0 in position 37: invalid start byte
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/opt/anaconda3/envs/fslpy/bin/fmriprep", line 8, in <module>
sys.exit(main())
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/fmriprep/cli/run.py", line 40, in main
parse_args()
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/fmriprep/cli/parser.py", line 878, in parse_args
config.from_dict({})
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/fmriprep/config.py", line 729, in from_dict
execution.load(settings, init=initialize('execution'), ignore=ignore)
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/fmriprep/config.py", line 240, in load
cls.init()
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/fmriprep/config.py", line 503, in init
cls._layout = BIDSLayout(
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/bids/layout/layout.py", line 177, in __init__
_indexer(self)
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/bids/layout/index.py", line 160, in __call__
self._index_metadata()
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/bids/layout/index.py", line 443, in _index_metadata
file_md.update(pl())
File "/opt/anaconda3/envs/fslpy/lib/python3.10/site-packages/bids/layout/index.py", line 333, in load_json
raise OSError(
OSError: Error occurred while trying to decode JSON from file <BIDSJSONFile filename='/Volumes/BENO_ALBE/prova/MS_FATIGUE_BIDS_format/._task-rest_bold.json'