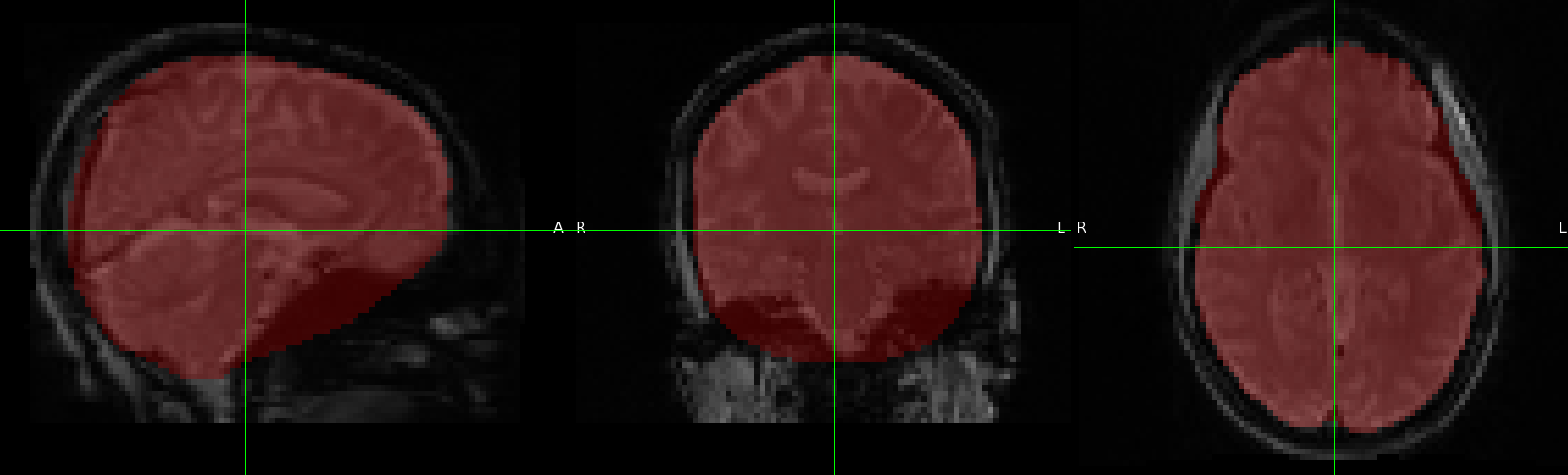
Hi all,
I am encountering an issue with the BOLD brain mask being generated by fmriprep including too much non-brain tissue. The pipeline we’re using goes on to reference this mask in subsequent calls and is resulting in a very noisy and inaccurate summary of components and signals, particularly in the area shown inferior to the frontal/temporal lobes.
Is there any way to adjust the singularity command to create a more appropriate mask for this data?
Our original image is being acquired as a multi-echo fMRI, and we are running the HCP Preprocessing pipeline/FreeSurfer prior to fmriprep.
We are running fmriprep-23.2.3 and here is the singularity call.
singularity run --cleanenv \
--no-home \
-B ${jobTmpDir}:/tmp \
-B "/project/***/templateflow:/templateflow" \
-B "/appl/freesurfer-7.1.1:/freesurfer" \
-B ${inputdir}:/data/input \
-B ${outputdir}:/data/output \
-B /project/***/***/fmriprep/prerun_output/$1:/fssubdir \
/project/***/singularity_containers/fmriprep-23.2.3.simg \
/data/input /data/output participant --skull-strip-template OASIS30ANTs --fs-license-file /freesurfer/license.txt \
--fs-subjects-dir /fssubdir/sourcedata/freesurfer \
--output-spaces fsaverage T1w fsnative fsLR MNI152NLin6Asym:res-2 --cifti-output 91k \
--bold2t1w-dof 6 \
--dvars-spike-threshold 1.5 \
--fd-spike-threshold 0.5 \
--ignore slicetiming \
--me-output-echos \
--notrack --nthreads 16 --omp-nthreads 15 --work-dir ${SINGULARITYENV_TMPDIR} --verbose --participant-label $1
Thanks in advance.