Summary of what happened:
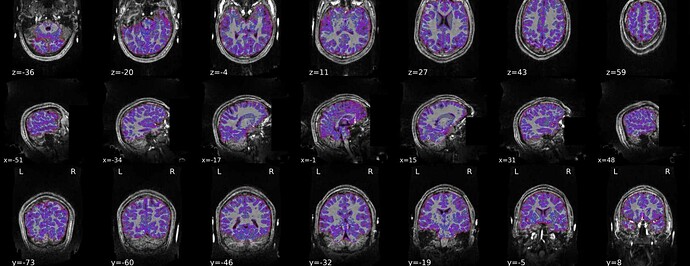
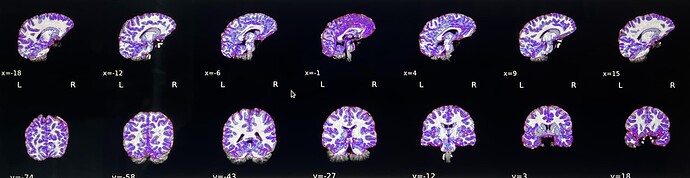
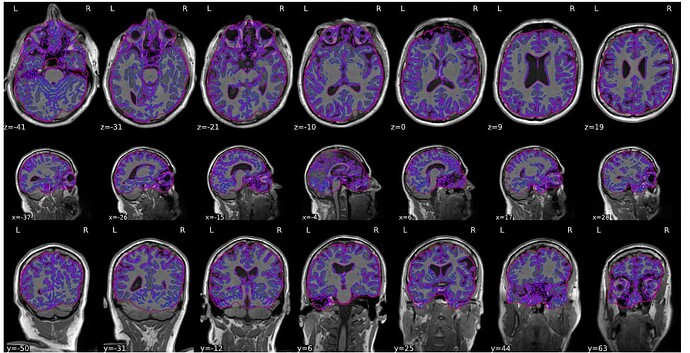
We’re encountering a persistent issue with mask generation during fMRIPrep processing, which results in the cerebellum being cut off and a stretched T1w-to-MNI normalization (see video here). After troubleshooting, we identified that the problem might be related to ANTs. We attempted brain extraction using optiBET, which successfully retained the cerebellum (see video here).
Following this, we tried providing the skull-stripped T1 image from optiBET directly to fMRIPrep as the input file, hoping it would simplify segmentation. However, the problem persisted, with the cerebellum still being cut off in the generated masks.
Our next step was to use the output masks from optiBET in fMRIPrep via the --derivatives flag. Unfortunately, we’re constrained to fMRIPrep version 20.0.0 on our cluster, and it seems this flag wasn’t introduced until version 22.3.0.
If anyone has suggestions or alternative approaches, we would greatly appreciate the help—we’ve been stuck on this for weeks!
Command used (and if a helper script was used, a link to the helper script or the command generated):
I run the following in the terminal which calls on the scripts linked below:
python iteachmri.py --json data_iteachmri.json --steps fmriprep --sub {X}
The code that executes the command (found in the helper script) is as follows:
fmriprepCommand = '"singularity run -B /ncf /n/sw/ncf/containers/' + fmriprep_module + '/fmriprep.sif ' + BIDSDir + ' ' + outDir + ' participant --participant-label ' + sub + ' --fs-license-file ${FREESURFER_HOME}/license.txt --nthreads ' + str(nThreads) + ' --mem ' + str(mem) + ' --stop-on-first-crash -w ' + workingDir + ' --output-spaces ' + spaces + ' --verbose --skull-strip-t1w skip --derivatives ' + outDir + '/' + sub + '"'
Link to runner script
Link to helper script
Version:
20.0.0
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity container on university cluster