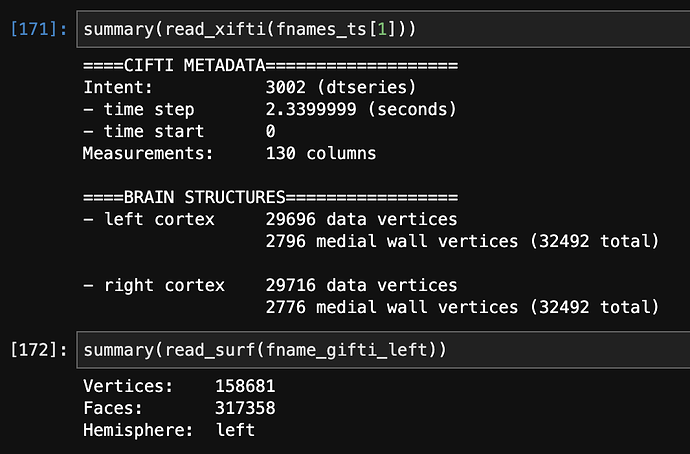
I definitely could have explained that better. As an example, in following some of the CIFTI examples in this tutorial, I am trying to run the following to plot the CIFTI data overlaid the subject surface (with paths replaced by placeholders):
cifti = nb.load('/fmriprep/sub-XXX/func/sub-XXX_task-XXX_space-fsLR_den-91k_bold.dtseries.nii')
cifti_data = cifti.get_fdata(dtype=np.float32)
cifti_hdr = cifti.header
nifti_hdr = cifti.nifti_header
axes = [cifti_hdr.get_axis(i) for i in range(cifti.ndim)]
_ = nlp.plot_surf("/fmriprep/sub-XXX/anat/sub-XXX_hemi-L_midthickness.surf.gii",
surf_data_from_cifti(cifti_data, axes[1], 'CIFTI_STRUCTURE_CORTEX_LEFT').mean(axis=1),
cmap='plasma')
I get the following error:
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/tmp/ipykernel_31980/3078201193.py in <module>
----> 1 _ = nlp.plot_surf("/om4/group/gablab/data/ds003126/derivatives/fmriprep/sub-047EPKL011005/ses-1/anat/sub-047EPKL011005_ses-1_hemi-L_midthickness.surf.gii",
2 surf_data_from_cifti(cifti_data, axes[1], 'CIFTI_STRUCTURE_CORTEX_LEFT').mean(axis=1),
3 cmap='plasma')
/om2/user/smeisler/anaconda3/envs/nipype/lib/python3.8/site-packages/nilearn/plotting/surf_plotting.py in plot_surf(surf_mesh, surf_map, bg_map, hemi, view, cmap, colorbar, avg_method, threshold, alpha, bg_on_data, darkness, vmin, vmax, cbar_vmin, cbar_vmax, cbar_tick_format, title, output_file, axes, figure, **kwargs)
231 '%i dimensions' % surf_map_data.ndim)
232 if surf_map_data.shape[0] != coords.shape[0]:
--> 233 raise ValueError('The surf_map does not have the same number '
234 'of vertices as the mesh.')
235
ValueError: The surf_map does not have the same number of vertices as the mesh.
Here are the numbers of vertices of each file from the ciftiTools package in R (the first part is from the functional data, second from the anatomical surfaces):