Hi everyone, I’m new to fmriprep and am working on my first project. I ran the following bash script:
#User inputs:
bids_root_dir=/home/11344938/MOUS/MOUS-subset/MOUS_BIDS/
subj=(sub-V1005 sub-V1016 sub-V1017 sub-V1029 sub-V1035 sub-V1046 sub-V1053 sub-V1059 sub-V1077 sub-V1083 sub-V1089 sub-V1095 sub-V1101 sub-V1107 sub-V1114)
for subj_idx in "${!subj[@]}"; do
echo preprocessing subject ${subj[$subj_idx]}
mkdir -p /home/11344938/MOUS/MOUS-subset/derrivatives/${subj[$subj_idx]}
fmriprep-docker $bids_root_dir $bids_root_dir \
participant \
--participant-label ${subj[$subj_idx]} \
--skip-bids-validation \
--fs-license-file /home/11344938/MOUS/MOUS-subset/MOUS_BIDS/fs_license.txt \
--output-spaces MNI152NLin2009cAsym:res-2 \
--stop-on-first-crash \
-w /home/11344938/MOUS/MOUS-subset/derrivatives/${subj[$subj_idx]}
done
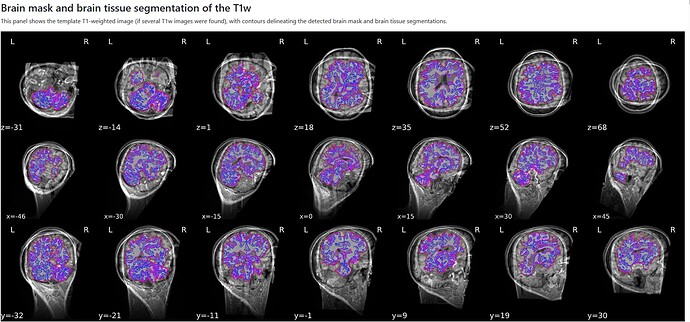
However, when inspecting the HTML file, the Brain mask and brain tissue segmentation of the T1w were misaligned entirely, as seen in the image below.
This happened for all the participants even though they ran without any errors. Would anyone know what’s going on here?
fmriprep-docker $bids_root_dir $bids_root_dir ...
You can’t use the input directory as the output directory. This is practically guaranteed to break something. Assuming that this isn’t the cause of the error, reading the docs:
The anatomical sub-workflow begins by constructing an average image by conforming all found T1w images to RAS orientation and a common voxel size, and, in the case of multiple images, averages them into a single reference template (see Longitudinal processing).
In the case of multiple T1w images (across sessions and/or runs), T1w images are merged into a single template image using FreeSurfer’s mri_robust_template.
It would seem you have two input T1w images, and at least one of them has an affine that is very far from accurate. If you simply load the two up in a viewer like Freeview, what do you see?
Thanks for your quick response! I looked at the two t1w images and this lines up very closely to what I’m seeing in the output HTML file. I think I made a mistake in my .bidsignore file. I will try and fix this as well as define another output directory and report if this indeed fixes the issue. Thanks again.
I strongly advise against using .bidsignore files for anything except encoding data that is not yet encodable in BIDS. It is only a validator concept intended for that purpose, and most tools will not respect it, including fMRIPrep. This is by design, not a “to-be-implemented” feature. There’s an extensive discussion on the topic here, if you’re interested: Standardize .bidsignore · Issue #131 · bids-standard/bids-specification · GitHub
If you have versions of your data that are unfit for consumption but you want to keep around for provenance reasons, the sourcedata/ directory is good for that.