Hi all,
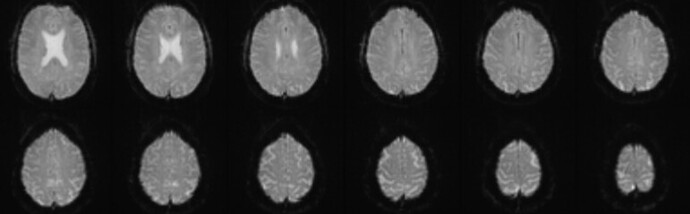
I’m using fMRIPrep to preprocess fMRI done with a protocol that didn’t fully cover the superior part of the brain. See the last slices of an example DCM volume.
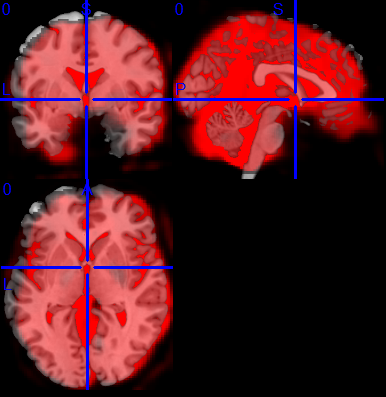
fMRIPrep, by default on this data, chose BBR registration, and probably because the superior border was by definition impossible to match, the coreg was bad. How bad? as below…
Running with –force-no-bbr however, did yield fair results, as shown below:
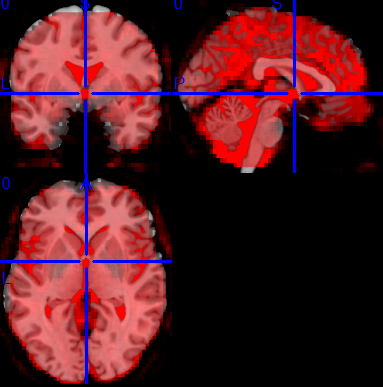
Seemingly, a happy ending.- so what is the question?
Basically, I’m not content of the idea of running two methods on data that will inevitably end with a poor fit and see which will somehow get away with it. And consequently, I’m wondering whether there’s a way to run BBR but inform it not to penalize border with, say Z values greater than threshold - or at least - have a loss that is asymmetrical - to penalize only BOLD borders that are not met by T1 borders but not the other way round - or anything alike…
Is something like that possible?
Thanks!