Summary of what happened:
I am interested in using the MIITRA 1 mm template for my older adult study. However, my code keeps failing to run. I went through the Template Flow directions as stated in the fmriprep website and tried troubleshooting based on previous forum discussions, but it still fails. I’d appreciate any advice or insight, thank you!
Note: if I take out the line that mounts templateflow and change the output space to T1w, my code runs fine.
Command used (and if a helper script was used, a link to the helper script or the command generated):
docker run -it --rm \
-v [path to data]:/data:ro \
-v [path to freesurfer license]:/opt/freesurfer/license.txt \
-v [path to output folder]/:/out \
-v [path to CustomTemplateFlow]:/home/fmriprep/.cache/templateflow \
nipreps/fmriprep:23.2.1 \
/data /out participant \
--skip_bids_validation \
--fs-no-reconall \
--ignore slicetiming \
--fd-spike-threshold 0.8 \
--dvars-spike-threshold 1.8 \
--nprocs 10 \
--participant_label sub-002 \
--output-spaces /tpl-MIITRA1mm
Version:
23.2.1
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Relevant log outputs (up to 20 lines):
fmriprep: error: One or more participant labels were not found in the BIDS directory: .
Note: this log output doesn't make sense to me because my code runs fine when using a pre-existing template, so I don't think it's a BIDS issue.
Screenshots / relevant information:
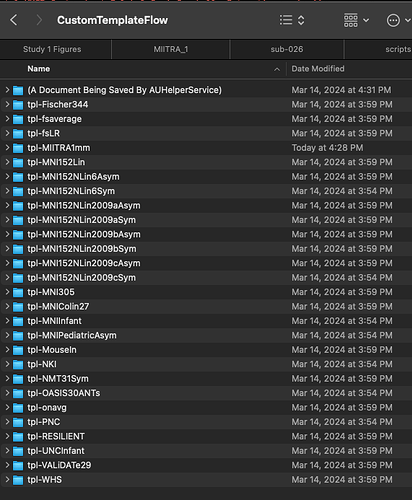
My CustomTemplateFlow folder is organized as follows:
CustomTemplateFlow /
tpl-MIITRA1mm /
template_description.json
tpl-MIITRA1mm_desc-brain_mask.nii.gz
tpl-MIITRA1mm_T1w.nii.gz
Trouble-shooting I’ve attempted:
- Tried different versions of fMRIprep
- Tried -e TEMPLATEFLOW_HOME=‘/CustomTemplateFlow’
- Tried specifying my subject as 002 instead of sub-002
- Tried changing the output space to “/MIITRA1mm” and “MIITRA1mm”