Hi @Steven
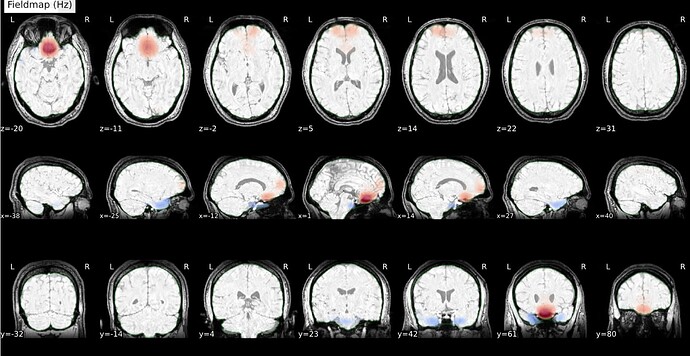
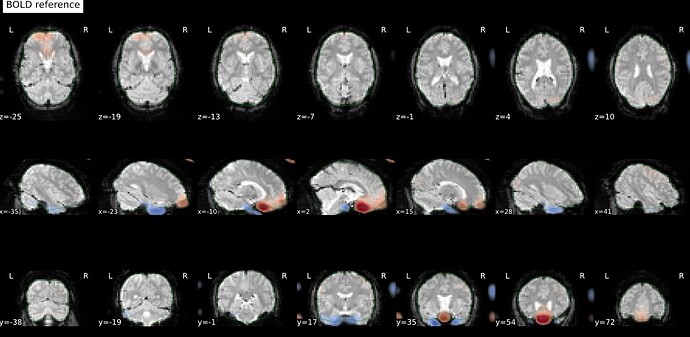
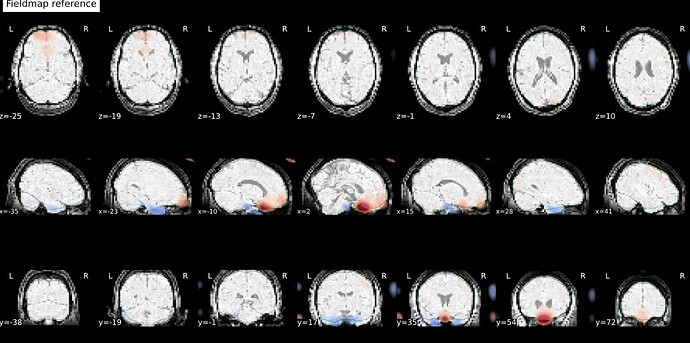
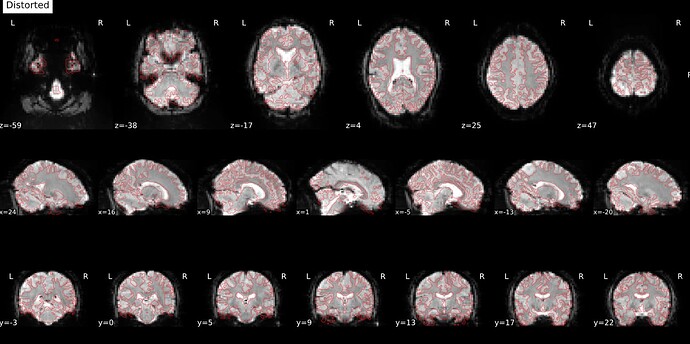
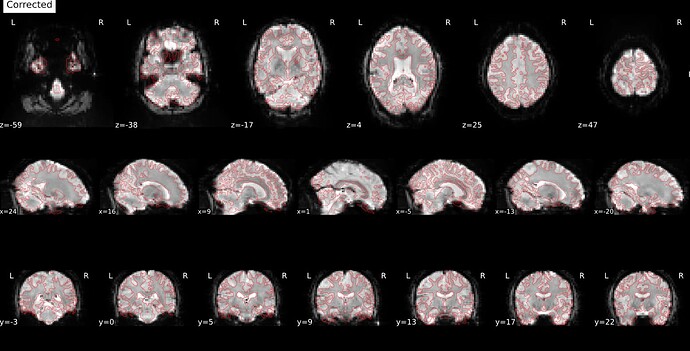
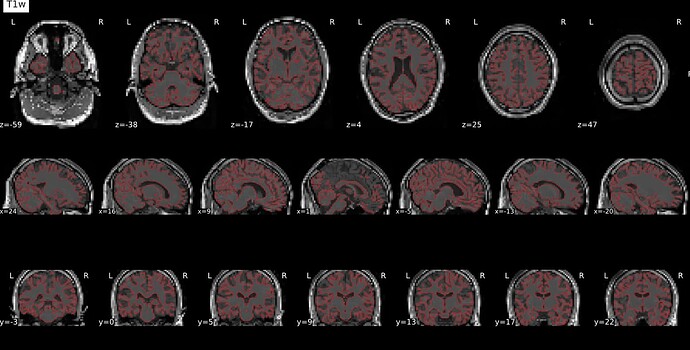
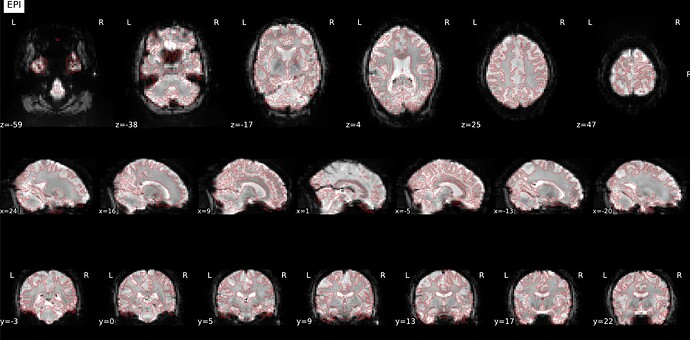
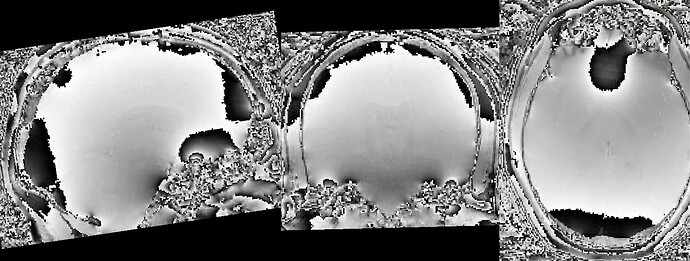
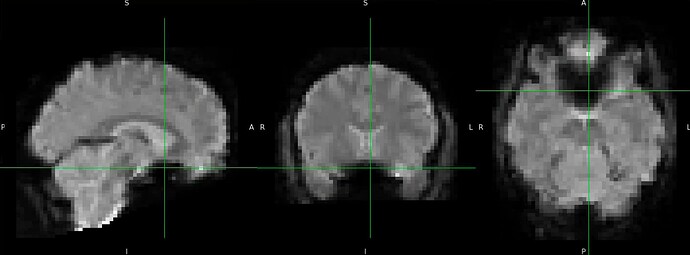
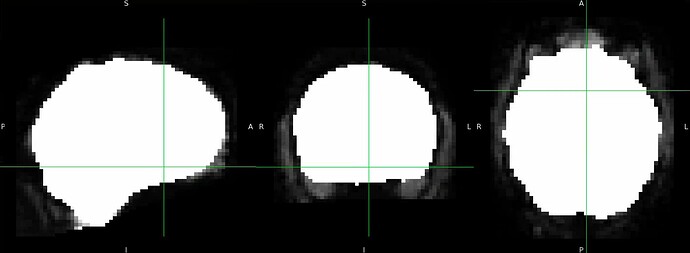
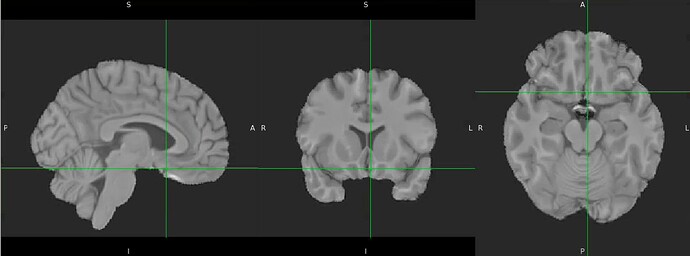
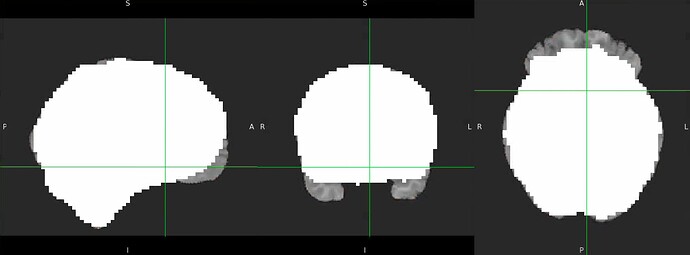
Thanks. I didn’t have this item specified in the func json but have added it. This hasn’t changed the resulting map. I have copied the full json below. I have attached the T1w image and BOLD image in MNI space and their respective masks to illustrate the failed normalization of the BOLD image, either due to error with distortion correction or coregistration with the T1w, as the T1w normalization is perfect.
Thanks, Luke
sub-031_task-rest_run-1_bold.json
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 123.256375,
"Manufacturer": "Siemens",
"InstitutionalDepartmentName": "Department",
"InstitutionAddress": "Du Cane Road StreetNo,London,District,GB,W12 0HS",
"DeviceSerialNumber": "166001",
"BodyPartExamined": "HEAD",
"PatientPosition": "HFS",
"ProcedureStepDescription": "MRI Head",
"SoftwareVersions": "syngo MR E11",
"MRAcquisitionType": "2D",
"SeriesDescription": "BOLD - Resting State",
"ProtocolName": "BOLD - Resting State",
"ScanningSequence": "EP",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"SequenceName": "*epfid2d1_64",
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"ND",
"NORM",
"MOSAIC"
],
"NonlinearGradientCorrection": false,
"SeriesNumber": 41,
"AcquisitionTime": "12:46:14.277500",
"AcquisitionNumber": 1,
"SliceThickness": 3,
"SpacingBetweenSlices": 3,
"SAR": 0.10801,
"EchoTime": 0.03,
"RepetitionTime": 2.5,
"FlipAngle": 90,
"PartialFourier": 1,
"BaseResolution": 64,
"ShimSetting": [
8824,
-8129,
-129,
302,
-16,
-154,
197,
7
],
"TxRefAmp": 279.714,
"PhaseResolution": 1,
"VendorReportedEchoSpacing": 0.00071,
"ReceiveCoilName": "HeadNeck_64",
"ReceiveCoilActiveElements": "HC3-6",
"PulseSequenceDetails": "%SiemensSeq%\\ep2d_pace",
"RefLinesPE": 24,
"CoilCombinationMethod": "Sum of Squares",
"ConsistencyInfo": "N4_VE11C_LATEST_20160120",
"MatrixCoilMode": "GRAPPA",
"PercentPhaseFOV": 100,
"PercentSampling": 100,
"EchoTrainLength": 31,
"PhaseEncodingSteps": 63,
"AcquisitionMatrixPE": 64,
"ReconMatrixPE": 64,
"BandwidthPerPixelPhaseEncode": 44.014,
"ParallelReductionFactorInPlane": 2,
"EffectiveEchoSpacing": 0.000355001,
"DerivedVendorReportedEchoSpacing": 0.000710001,
"TotalReadoutTime": 0.022365,
"PixelBandwidth": 1595,
"DwellTime": 4.9e-06,
"PhaseEncodingDirection": "j-",
"SliceTiming": [
0,
0.06,
0.1175,
0.1775,
0.2375,
0.295,
0.355,
0.4125,
0.4725,
0.5325,
0.59,
0.65,
0.7075,
0.7675,
0.8275,
0.885,
0.945,
1.005,
1.0625,
1.1225,
1.18,
1.24,
1.3,
1.3575,
1.4175,
1.475,
1.535,
1.595,
1.6525,
1.7125,
1.7725,
1.83,
1.89,
1.9475,
2.0075,
2.0675,
2.125,
2.185,
2.2425,
2.3025,
2.3625,
2.42
],
"ImageOrientationPatientDICOM": [
0.997087,
-0.040086,
0.0648888,
0.0697231,
0.823941,
-0.56237
],
"ImageOrientationText": "Tra>Cor(-34.4)>Sag(1.8)",
"InPlanePhaseEncodingDirectionDICOM": "COL",
"BidsGuess": [
"func",
"_acq-epfid2p2_dir-AP_run-41_bold"
],
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20240202",
"TaskName": "rest",
"B0FieldSource": "fmap0"
}
BOLD in MNI space
BOLD mask in MNI space
T1w in MNI space
T1w with the BOLD mask ontop in MNI space
Demonstrates the misalignment.