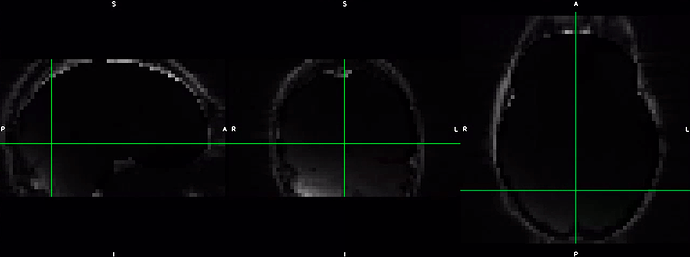
Dear fmriprep experts,
I am finding very strange results following the N4 bias field correction to the ref_bold.nii.gz file of one of my functional runs for one subject. Below is the ref_bold_corrected.nii.gz file for that functional run.
This cascades throughout the remaining pipeline providing unusable data.
This thread suggests using --force-bbr when calling fmriprep, but that failed to solve this problem for me.
Outside of fmriprep, I called N4BiasFieldCorrection using the default fmriprep arguments with the addition of --shrink-factor 3 . That manages to produce a reasonable image.
Is it possible to run the N4BiasFieldCorrection inside the fmriprep call with the --shrink-factor 3 included?