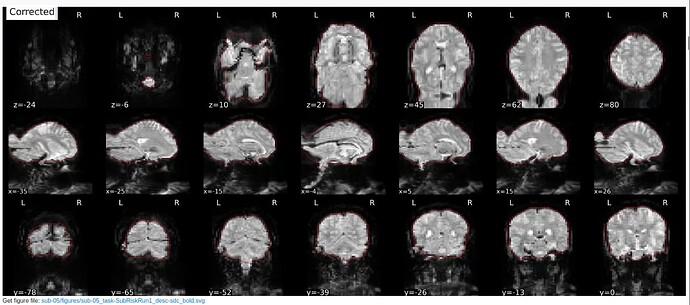
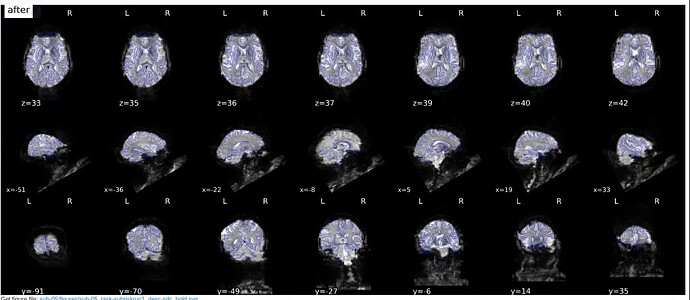
Summary of what happened:
Hello,
I’m using fmriprep with docker and I’ve been having problems with SDC using phasediff.
In another thread related to similar SDC problems (Fieldmap correction gone wrong - #18 by effigies) someone recommended to use the pre-release 23.0.2a2, but when I run it I always get the same error.
Does anyone know what the problem might be in any of these cases?
Thank you in advance!
Command used (and if a helper script was used, a link to the helper script or the command generated):
docker run -ti --rm \
-v /media/patricia/data/fmriprep_data:/data:ro \
-v /media/patricia/data/output_fmriprep:/out \
-v /home/patricia/freesurfer/license.txt:/opt/freesurfer/license.txt:ro \
nipreps/fmriprep:23.2.0a2 \
/data /out/fmriprep \
participant --participant-label sub-05 \
Version:
23.2.0a2
Environment (Docker, Singularity, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
File Path: Task scans should have a corresponding events.tsv file. If this is a resting state scan you can ignore this warning or rename the task to include the word "rest".
Type: Warning
File: sub-05_task-SubRiskRun1_bold.nii.gz
Location: f/sub-05/func/sub-05_task-SubRiskRun1_bold.nii.gz
Reason: Task scans should have a corresponding events.tsv file. It can be included one of the following locations: /events.tsv, /task-SubRiskRun1_events.tsv, /sub-05/sub-05_events.tsv, /sub-05/sub-05_task-SubRiskRun1_events.tsv, /sub-05/func/sub-05_events.tsv, /sub-05/func/sub-05_task-SubRiskRun1_events.tsv
Type: Warning
File: sub-05_task-SubRiskRun2_bold.nii.gz
Location: f/sub-05/func/sub-05_task-SubRiskRun2_bold.nii.gz
Reason: Task scans should have a corresponding events.tsv file. It can be included one of the following locations: /events.tsv, /task-SubRiskRun2_events.tsv, /sub-05/sub-05_events.tsv, /sub-05/sub-05_task-SubRiskRun2_events.tsv, /sub-05/func/sub-05_events.tsv, /sub-05/func/sub-05_task-SubRiskRun2_events.tsv
Type: Warning
File: sub-05_task-SubRiskRun3_bold.nii.gz
Location: f/sub-05/func/sub-05_task-SubRiskRun3_bold.nii.gz
Reason: Task scans should have a corresponding events.tsv file. It can be included one of the following locations: /events.tsv, /task-SubRiskRun3_events.tsv, /sub-05/sub-05_events.tsv, /sub-05/sub-05_task-SubRiskRun3_events.tsv, /sub-05/func/sub-05_events.tsv, /sub-05/func/sub-05_task-SubRiskRun3_events.tsv
Type: Warning
File: sub-05_task-SubRiskRun4_bold.nii.gz
Location: f/sub-05/func/sub-05_task-SubRiskRun4_bold.nii.gz
Reason: Task scans should have a corresponding events.tsv file. It can be included one of the following locations: /events.tsv, /task-SubRiskRun4_events.tsv, /sub-05/sub-05_events.tsv, /sub-05/sub-05_task-SubRiskRun4_events.tsv, /sub-05/func/sub-05_events.tsv, /sub-05/func/sub-05_task-SubRiskRun4_events.tsv
======================================================
File Path: The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered based on the presence of only one author field. Please ignore if all contributors are already properly listed.
Type: Warning
======================================================
Relevant log outputs (up to 20 lines):
Node: fmriprep_23_2_wf.sub_05_wf.bold_task_subriskrun1_wf.bold_native_wf.boldref_fmap
Working directory: /tmp/work/fmriprep_23_2_wf/sub_05_wf/bold_task_subriskrun1_wf/bold_native_wf/boldref_fmap
Node inputs:
fmap_ref_file =
in_coeffs =
inverse = [True]
target_ref_file =
transforms =
Traceback (most recent call last):
File “/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py”, line 67, in run_node
result[“result”] = node.run(updatehash=updatehash)
File “/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py”, line 527, in run
result = self._run_interface(execute=True)
File “/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py”, line 645, in _run_interface
return self._run_command(execute)
File “/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py”, line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node boldref_fmap.
Traceback:
Traceback (most recent call last):
File “/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py”, line 397, in run
runtime = self._run_interface(runtime)
File “/opt/conda/envs/fmriprep/lib/python3.10/site-packages/fmriprep/interfaces/resampling.py”, line 169, in _run_interface
fieldmap = reconstruct_fieldmap(
File “/opt/conda/envs/fmriprep/lib/python3.10/site-packages/fmriprep/interfaces/resampling.py”, line 661, in reconstruct_fieldmap
raise ValueError(‘Reference passed is not aligned with spline grids’)
ValueError: Reference passed is not aligned with spline grids
Screenshots / relevant information:
I first tried version 23.1.4 and later version 20.2.3 - you can see how both output look in the images below with both versions on the same subject.
With v. 23.1.4
With v.20.2.3