Summary of what happened:
I did coregistration and reslice using spm, and then used the spm-processed files to do fmriprep but failed with error (this Node: fmriprep_24_0_wf.sub_1439_wf.fmap_preproc_wf.wf_auto_00000.magnitude_wf.brainextraction_wf.clipper_pre)
I can’t figure out which part of the whole process is wrong.
Command used (and if a helper script was used, a link to the helper script or the command generated):
#spm code:
import os
import gzip
import shutil
from nipype.interfaces.spm import Coregister
from glob import glob
# Paths
base_dir = "/Users/sap/Desktop/UTO_SPM_reslice"
# Unzip .nii.gz files
def unzip_files(folder):
gz_files = glob(os.path.join(folder, "*.nii.gz"))
for gz_file in gz_files:
nii_file = gz_file.replace(".gz", "")
with gzip.open(gz_file, "rb") as f_in, open(nii_file, "wb") as f_out:
shutil.copyfileobj(f_in, f_out)
# Run SPM coregistration and reslicing
def spm_coregistration(sub_id):
sub_path = os.path.join(base_dir, f"sub-{sub_id}")
func_file = os.path.join(sub_path, "func", f"sub-{sub_id}_task-rest_bold.nii")
fmap_folder = os.path.join(sub_path, "fmap")
magnitude1 = os.path.join(fmap_folder, f"sub-{sub_id}_magnitude1.nii")
magnitude2 = os.path.join(fmap_folder, f"sub-{sub_id}_magnitude2.nii")
phasediff = os.path.join(fmap_folder, f"sub-{sub_id}_phasediff.nii")
# Configure SPM Coregistration
coreg = Coregister()
coreg.inputs.target = func_file
coreg.inputs.source = magnitude1
coreg.inputs.apply_to_files = [magnitude2, phasediff]
coreg.inputs.cost_function = "nmi"
coreg.inputs.separation = [4, 2]
coreg.inputs.tolerance = [0.02] * 3 + [0.001] * 3 + [0.01] * 3 + [0.001] * 3
coreg.inputs.fwhm = [7, 7]
coreg.inputs.write_interp = 4
coreg.inputs.write_wrap = [0, 0, 0]
coreg.inputs.jobtype = "estwrite"
coreg.run()
# Clean up and rename files
def cleanup_and_rename(sub_id):
sub_path = os.path.join(base_dir, f"sub-{sub_id}")
fmap_folder = os.path.join(sub_path, "fmap")
func_folder = os.path.join(sub_path, "func")
# For fmap folder
# Step 3: Delete original .nii.gz files
original_gz_files = glob(os.path.join(fmap_folder, "sub-*.nii.gz"))
for gz_file in original_gz_files:
os.remove(gz_file)
# Step 4: Rename and compress rsub- files
fmap_files = glob(os.path.join(fmap_folder, "*.nii"))
for file in fmap_files:
if "rsub-" in file: # Resliced files
new_name = file.replace("rsub-", "sub-")
os.rename(file, new_name)
# Compress back to .nii.gz
with open(new_name, "rb") as f_in, gzip.open(f"{new_name}.gz", "wb") as f_out:
shutil.copyfileobj(f_in, f_out)
os.remove(new_name) # Remove .nii
# For func folder
func_files = glob(os.path.join(func_folder, "*.nii"))
for file in func_files:
os.remove(file) # Delete .nii files
# Main loop for all subjects
for sub_folder in sorted(glob(os.path.join(base_dir, "sub-*"))):
sub_id = os.path.basename(sub_folder).split("-")[1]
print(f"Processing subject {sub_id}...")
# Step 1: Unzip files
unzip_files(os.path.join(sub_folder, "func"))
unzip_files(os.path.join(sub_folder, "fmap"))
# Step 2: Run SPM coregistration
spm_coregistration(sub_id)
# Step 3: Clean up and rename files
cleanup_and_rename(sub_id)
print("All subjects processed successfully.")
# fmriprep code
#!/bin/bash
# Input arguments
subject=$1 # Subject ID
FMRIPREP_VER=24.0.1
bids_root_dir=/Users/sap/Desktop/UTO_SPM_reslice
nthreads=$2 # Number of threads
# Notify that fMRIPrep is starting for this subject
echo "Starting fMRIPrep for $subject..."
# Run fmriprep
fmriprep-docker $bids_root_dir/sourcedata $bids_root_dir/derivatives/fmriprep \
participant \
--participant-label $subject \
--fs-license-file /Users/sap/Desktop/derivatives/license.txt \
--nthreads $nthreads \
--stop-on-first-crash \
--mem_mb 15000 \
--output-spaces MNI152NLin2009cAsym:res-2 T1w \
--return-all-components \
--image nipreps/fmriprep:$FMRIPREP_VER \
--no-tty
# Notify when fMRIPrep is done for this subject
echo "Finished fMRIPrep for $subject!"
Version:
fmriprep 24.0.1
spm12
Environment (Docker, Singularity / Apptainer, custom installation):
docker
Data formatted according to a validatable standard? Please provide the output of the validator:
./sourcedata
├── dataset_description.json
├── sub-1439
│ ├── anat
│ │ ├── sub-1439_T1w.json
│ │ └── sub-1439_T1w.nii.gz
│ ├── fmap
│ │ ├── sub-1439_magnitude1.json
│ │ ├── sub-1439_magnitude1.nii.gz
│ │ ├── sub-1439_magnitude2.json
│ │ ├── sub-1439_magnitude2.nii.gz
│ │ ├── sub-1439_phasediff.json
│ │ └── sub-1439_phasediff.nii.gz
│ └── func
│ ├── sub-1439_task-rest_bold.json
│ └── sub-1439_task-rest_bold.nii.gz
├── sub-1440
│ ├── anat
│ │ ├── sub-1440_T1w.json
│ │ └── sub-1440_T1w.nii.gz
│ ├── fmap
│ │ ├── sub-1440_magnitude1.json
│ │ ├── sub-1440_magnitude1.nii.gz
│ │ ├── sub-1440_magnitude2.json
│ │ ├── sub-1440_magnitude2.nii.gz
│ │ ├── sub-1440_phasediff.json
│ │ └── sub-1440_phasediff.nii.gz
│ └── func
│ ├── sub-1440_task-rest_bold.json
│ └── sub-1440_task-rest_bold.nii.gz
└── sub-1458
├── anat
│ ├── sub-1458_T1w.json
│ └── sub-1458_T1w.nii.gz
├── fmap
│ ├── sub-1458_magnitude1.json
│ ├── sub-1458_magnitude1.nii.gz
│ ├── sub-1458_magnitude2.json
│ ├── sub-1458_magnitude2.nii.gz
│ ├── sub-1458_phasediff.json
│ └── sub-1458_phasediff.nii.gz
└── func
├── sub-1458_task-rest_bold.json
└── sub-1458_task-rest_bold.nii.gz
13 directories, 31 files
Relevant log outputs (up to 20 lines):
fmriprep crash report:
Node: fmriprep_24_0_wf.sub_1439_wf.fmap_preproc_wf.wf_auto_00000.magnitude_wf.brainextraction_wf.clipper_pre
Working directory: /tmp/work/fmriprep_24_0_wf/sub_1439_wf/fmap_preproc_wf/wf_auto_00000/magnitude_wf/brainextraction_wf/clipper_pre
Node inputs:
dtype = int16
in_file = <undefined>
invert = False
nonnegative = True
p_max = 99.98
p_min = 35.0
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node clipper_pre.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/niworkflows/interfaces/nibabel.py", line 467, in _run_interface
self._results["out_file"] = _advanced_clip(
^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/niworkflows/interfaces/nibabel.py", line 705, in _advanced_clip
a_min = np.percentile(
^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/numpy/lib/function_base.py", line 4283, in percentile
return _quantile_unchecked(
^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/numpy/lib/function_base.py", line 4555, in _quantile_unchecked
return _ureduce(a,
^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/numpy/lib/function_base.py", line 3823, in _ureduce
r = func(a, **kwargs)
^^^^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/numpy/lib/function_base.py", line 4722, in _quantile_ureduce_func
result = _quantile(arr,
^^^^^^^^^^^^^^
File "/opt/conda/envs/fmriprep/lib/python3.11/site-packages/numpy/lib/function_base.py", line 4831, in _quantile
slices_having_nans = np.isnan(arr[-1, ...])
~~~^^^^^^^^^
IndexError: index -1 is out of bounds for axis 0 with size 0
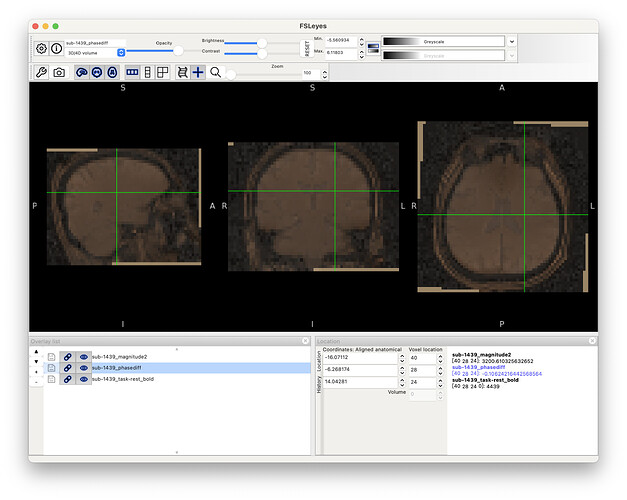
Screenshots / relevant information:
spm-processed files showed nice alignment: