Hi,
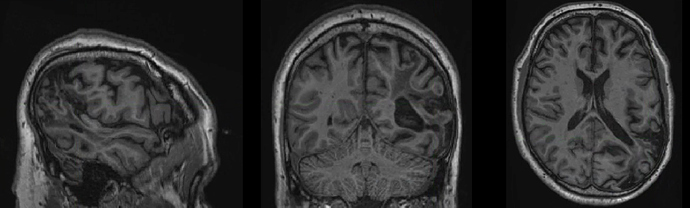
I’ve preprocessed data for several participants who have lesion masks. Unfortunately, the normalisation does not seem to be running well with the masks – the lesioned hemisphere seems to be bigger than the healthy one and is especially inflated around the lesion. I attach an example participant, this is the output in standard space MNI152NLin6Asym, it looks the same with the 2009 template and the same for the functional images.
Has anyone come accross this problem before? I assume other people have used fmriprep for data with lesion masks – did it work well for them? And does somebody know a solution to this?
Thanks!
Sandra
I wonder if anyone can help with this since I haven’t gotten any feedback? @effigies or @oesteban, would you have an idea? Thank you!
What is the outcome you’d expect?
(btw, there is this lingering feature implementation that you may want to take on - https://github.com/poldracklab/fmriprep/issues/1575)
Hi Sandra,
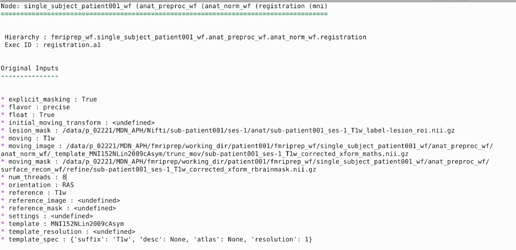
I have seen this before when running AntsRegistration without a lesion mask on data with a similar lesion, so I would start with double-checking if the lesion mask was actually used to run the registration (probably easiest to find out from the anat_preproc_wf/anat_norm_wf/_template_MNI152NLin2009cAsym/registration/_report/report.rst). If it did use the mask, could you then include an image that also has the mask in it?
Cheers,
Milanka
Hi Milanka,
Thank you for your reply! I just checked and yes, the lesion mask appears to have been included in the registration process as the output shows:
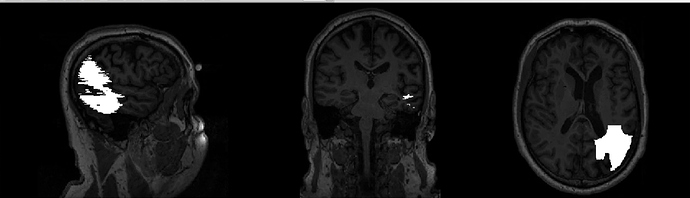
And here’s a picture of the anatomy and lesion mask before preprocessing:
When I look at the brain mask output in MNI space, it somehow doesn’t look to me as if it included the whole lesion:
What do you think?
Thanks,
Sandra
Thanks for the link. I’ve tried the toolbox with the enantiomorphic registration for these data and that worked well, I guess it would be a great addition to have this option in the pipeline.
Relating to my issue: The lesioned hemisphere is strongly inflated after preprocessing although the mask was used as input. I wonder why this happened since I would love to stick with fMRIPrep also with patient data.
Hi Sandra,
I would suggest running the registration with a more aggressive mask, especially masking the area in that wedge above the posterior temporal lobe. There is considerable warping of brain tissue extending towards that area judging from stretching of the ventricle. With masking that wedge, ANTs should then avoid warping healthy tissue into this area and which hopefully will give you a better result.
From looking at the brain mask, I’m guessing that you used recon-all and in that case the brain mask looks as expected. The brain mask is refined using the surface reconstruction and it this does not perform well in the lesioned area resulting in a gap in the brain mask.
Cheers,
Milanka
With our lesioned brains, I get similar brain masks where the whole lesion is not included. As Milanka mentioned the brain mask is refined using the surface reconstruction and this does not perform well in the lesioned area resulting in a gap in the brain mask. If you do not need FreeSurfer preprocessing and the bold signal sampled on the surface, you can use the --fs-no-reconall fmriprep argument to skip surface reconstruction. Using this argument, the brain masks include the whole lesion for our data.
Hi @milanka and @heffjos,
Thank you for your suggestion! I took out freesurfer and that really made a difference! The preprocessed images are now well aligned with the template. One more question though: After the preprocessing without freesurfer, the brain mask does not include the lesioned area anymore (see image below). Is that normal? I would have expected that the lesion would be included nonetheless.
And do you know if there is a a way in fmriprep to output a normalized version of the lesion mask as well or do you usually also use other tools for this?
Yes this is normal. fmriprep uses ANTs to register the T1w to a template and fmriprep uses an exclusion mask to mask the T1w image in ANTs rather than the brain mask. The exclusion mask has the same dimensions as the T1w and has value 1 at every voxel except at the lesion. For the lesion voxels, their value is set to 0.
Other tools are need for this, probably only ANTs. The transformation from T1w to template space is in the .h5 file in the anat directory (example: sub-001s01_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5). This topic might have more useful and detailed information how to use the .h5 files: Extracting individual transforms from composite .h5 files (fmriprep) I used the first example in the first post to transform a lesion mask from T1w to template space, and the results looked fine.
Thanks for the clarifications on the brain mask! This is really helpful!
And also thank you on your input on the transformation of the lesion mask – I’ve been using FSL in the past and will give ANTs a try for sure!
Best
Sandra
Hi heffjos,
Just wondering if you could please elaborate on this point a bit more. I am using fmriprep to pre-process a patient dataset and want to apply the T1 transformation matrix to MNI space to the lesion file. When you did this was the call something like…
antsApplyTransforms -d 3 --float 1 --verbose 1 -i sublesion.nii.gz -o sublesion.MNISpace.nii.gz -r MNI152NLin2009cAsym_res-01_desc-brain_T1w.nii -t sub_T1w_target-MNI152NLin2009cAsym_warp.h5 -n NearestNeighbor
I’m trying something like this and have had difficulty bringing the lesion into MNI space. I’m not entirely sure what’s going on and could use the help of others who have made it work.
Thanks!
Frank
Hi Sandra,
Just wondering if you ever identified a solution to bring your lesion into MNI space using the same T1 transformation matrix that was output by fmriprep. I asked a similar question to heffjos and/but figured I’d run it by you as well.
Thanks!
Frank