Hi fmriprep community!
Summary of what happened:
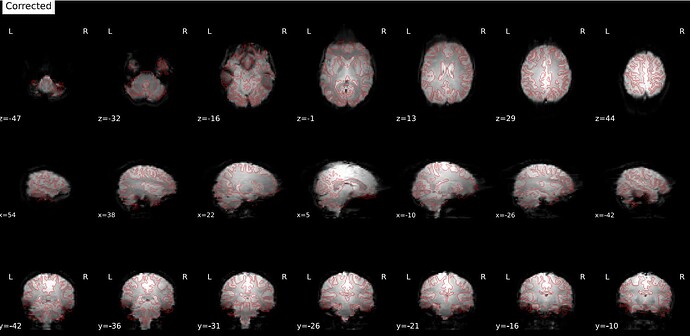
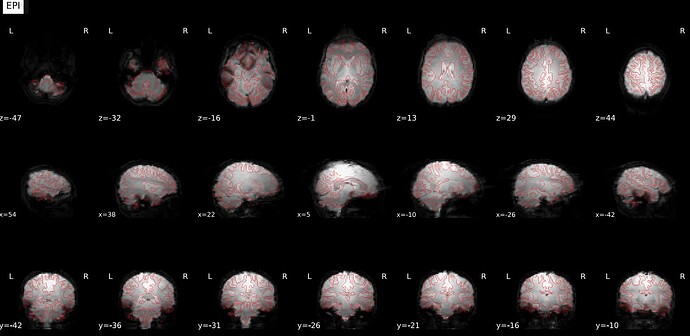
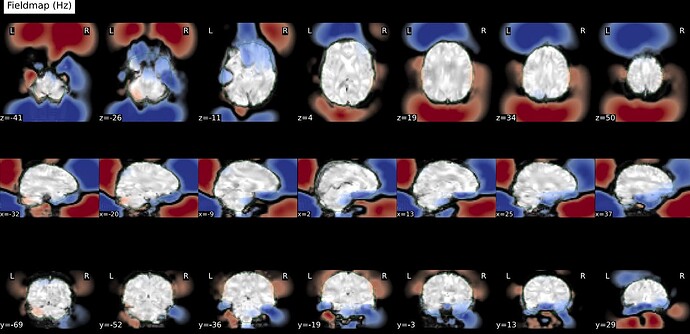
I ran fmriprep on five test subjects for one of my projects, but it seems the susceptibility distortion correction is introducing weird frontal and occipital artifacts (see screenshot below). Those artifacts create a misalignment in the frontal lobe when registering the BOLD data onto the anatomical space. I originally thought I got my phase encoding direction wrong. Still, after validation at the MRI console, it seems I got it correct (data comes from a Phillips 3T scanner, rs-fMRI PA with one reverse-encoded AP). Here are the other avenues I’ve tested without success:
- Use the
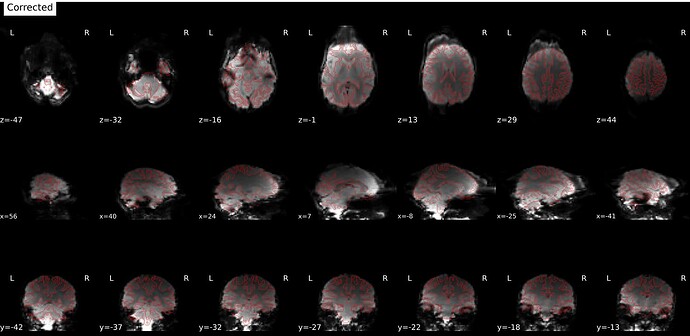
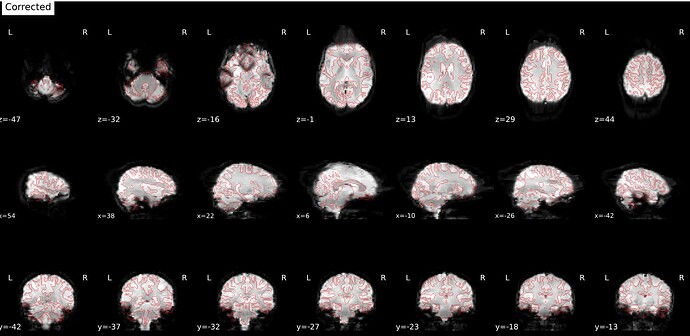
--ignore fmap_jacobianoption. It still produces the same artifact (it was actually worse). - Manually extracted a few volumes from the rs-fMRI sequence to create a distinct fmap with PA encoding. Same results.
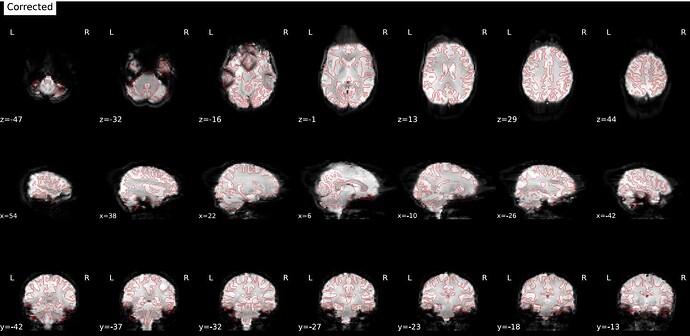
- Remove slice timing correction; still the same results.
Anyone encountered the same issue?
Command used (and if a helper script was used, a link to the helper script or the command generated):
#!/bin/bash
#
#SBATCH -J fmriprep
#SBATCH --time=48:00:00
#SBATCH -n 1
#SBATCH --cpus-per-task=16
#SBATCH --mem=31G
# Outputs ----------------------------------
#SBATCH -o log/%x-%A-%a.out
#SBATCH -e log/%x-%A-%a.err
#SBATCH --mail-user=Anthony.Gagnon7@usherbrooke.ca
#SBATCH --mail-type=ALL
# ------------------------------------------
BIDS_DIR="$STUDY/fMRI_data"
DERIVS_DIR="/derivatives/fmriprep-24.1.1"
LOCAL_FREESURFER_DIR="$STUDY/sourcedata/freesurfer/"
SCRATCH="$STUDY/temp/"
# Prepare some writeable bind-mount points.
TEMPLATEFLOW_HOST_HOME=$HOME/templateflow
FMRIPREP_HOST_CACHE=$HOME/.cache/fmriprep
mkdir -p ${TEMPLATEFLOW_HOST_HOME}
mkdir -p ${FMRIPREP_HOST_CACHE}
# Prepare derivatives folder
mkdir -p ${BIDS_DIR}/${DERIVS_DIR}
# Make sure FS_LICENSE is defined in the container.
export APPTAINERENV_FS_LICENSE=$STUDY/license.txt
# Designate a templateflow bind-mount point
export APPTAINERENV_TEMPLATEFLOW_HOME="/templateflow"
APPTAINER_CMD="apptainer run --cleanenv -B $BIDS_DIR:/data -B ${TEMPLATEFLOW_HOST_HOME}:${APPTAINERENV_TEMPLATEFLOW_HOME} -B $SCRATCH:/work -B ${LOCAL_FREESURFER_DIR}:/fsdir $STUDY/fmriprep_24.1.1-mp2.sif"
# Parse the participants.tsv file and extract one subject ID from the line corresponding to this SLURM task.
subject=$( sed -n -E "$((${SLURM_ARRAY_TASK_ID} + 1))s/sub-(\S*)\>.*/\1/gp" ${BIDS_DIR}/participants.tsv )
# Remove IsRunning files from FreeSurfer
find ${LOCAL_FREESURFER_DIR}/sub-${subject}-fs/sub-${subject} -name "*IsRunning*" -type f -delete
# Compose the command line
cmd="${APPTAINER_CMD} /data /data/${DERIVS_DIR} participant --participant-label $subject -w /work/ -v -t rest --omp-nthreads 8 --nthreads 16 --mem_mb 30000 --output-spaces MNI152NLin2009cAsym --cifti-output 91k --fs-subjects-dir /fsdir/sub-${subject}-fs"
# Setup done, run the command
echo Running task ${SLURM_ARRAY_TASK_ID}
echo Commandline: $cmd
eval $cmd
exitcode=$?
# Output results to a table
echo "sub-$subject ${SLURM_ARRAY_TASK_ID} $exitcode" \
>> ${SLURM_JOB_NAME}.${SLURM_ARRAY_JOB_ID}.tsv
echo Finished tasks ${SLURM_ARRAY_TASK_ID} with exit code $exitcode
exit $exitcode
Version:
v24.1.1
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer, but rebuild to a more portable build of ANTs (since I’m running it on an old HPC server). See Building fmriprep singularity/docker containers, CPU instruction level support · Issue #2437 · nipreps/fmriprep · GitHub.
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator@1.14.14
(node:19940) Warning: Closing directory handle on garbage collection
(Use `node --trace-warnings ...` to show where the warning was created)
1: [WARN] Task scans should have a corresponding events.tsv file. If this is a resting state scan you can ignore this warning or rename the task to include the word "rest". (code: 25 - EVENTS_TSV_MISSING)
./sub-1005/ses-baseline/func/sub-1005_ses-baseline_task-Simon_run-01_bold.nii.gz
./sub-1005/ses-baseline/func/sub-1005_ses-baseline_task-Simon_run-02_bold.nii.gz
./sub-1005/ses-baseline/func/sub-1005_ses-baseline_task-Simon_run-03_bold.nii.gz
./sub-1009/ses-baseline/func/sub-1009_ses-baseline_task-Simon_run-01_bold.nii.gz
./sub-1009/ses-baseline/func/sub-1009_ses-baseline_task-Simon_run-02_bold.nii.gz
./sub-1009/ses-baseline/func/sub-1009_ses-baseline_task-Simon_run-03_bold.nii.gz
./sub-1010/ses-baseline/func/sub-1010_ses-baseline_task-Simon_run-01_bold.nii.gz
./sub-1010/ses-baseline/func/sub-1010_ses-baseline_task-Simon_run-02_bold.nii.gz
./sub-1010/ses-baseline/func/sub-1010_ses-baseline_task-Simon_run-03_bold.nii.gz
./sub-1011/ses-baseline/func/sub-1011_ses-baseline_task-Simon_run-01_bold.nii.gz
... and 5 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=EVENTS_TSV_MISSING for existing conversations about this issue.
2: [WARN] NIfTI file's header field for unit information for x, y, z, and t dimensions empty or too short (code: 41 - NIFTI_UNIT)
./sub-1005/ses-baseline/fmap/sub-1005_ses-baseline_acq-func_dir-PA_run-01_epi.nii.gz
./sub-1005/ses-baseline/fmap/sub-1005_ses-baseline_acq-func_dir-PA_run-02_epi.nii.gz
./sub-1005/ses-baseline/fmap/sub-1005_ses-baseline_acq-func_dir-PA_run-03_epi.nii.gz
./sub-1005/ses-baseline/fmap/sub-1005_ses-baseline_acq-func_dir-PA_run-04_epi.nii.gz
./sub-1009/ses-baseline/fmap/sub-1009_ses-baseline_acq-func_dir-PA_run-01_epi.nii.gz
./sub-1009/ses-baseline/fmap/sub-1009_ses-baseline_acq-func_dir-PA_run-02_epi.nii.gz
./sub-1009/ses-baseline/fmap/sub-1009_ses-baseline_acq-func_dir-PA_run-03_epi.nii.gz
./sub-1009/ses-baseline/fmap/sub-1009_ses-baseline_acq-func_dir-PA_run-04_epi.nii.gz
./sub-1010/ses-baseline/fmap/sub-1010_ses-baseline_acq-func_dir-PA_run-01_epi.nii.gz
./sub-1010/ses-baseline/fmap/sub-1010_ses-baseline_acq-func_dir-PA_run-02_epi.nii.gz
... and 10 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=NIFTI_UNIT for existing conversations about this issue.
Summary: Available Tasks: Available Modalities:
165 Files, 1.8GB Simon's Task MRI
5 - Subjects Resting State (eyes open)
1 - Session
If you have any questions, please post on https://neurostars.org/tags/bids.
Relevant log outputs (up to 20 lines):
Screenshots / relevant information:
Original try (PA rsfMRI with one reverse-encoded AP):
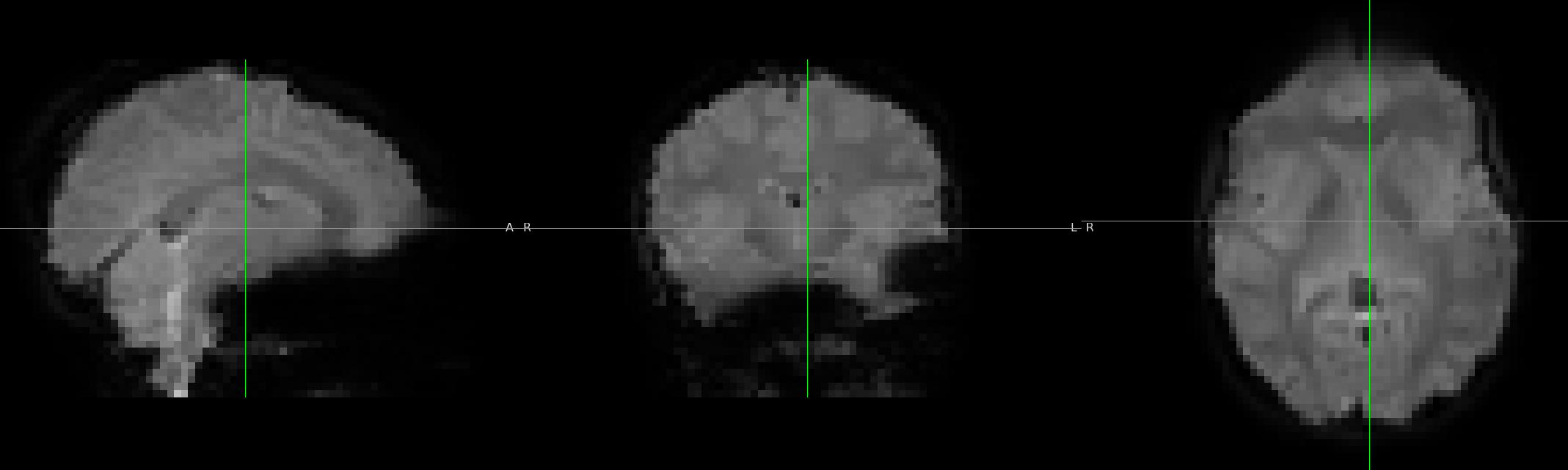
No jacobian:
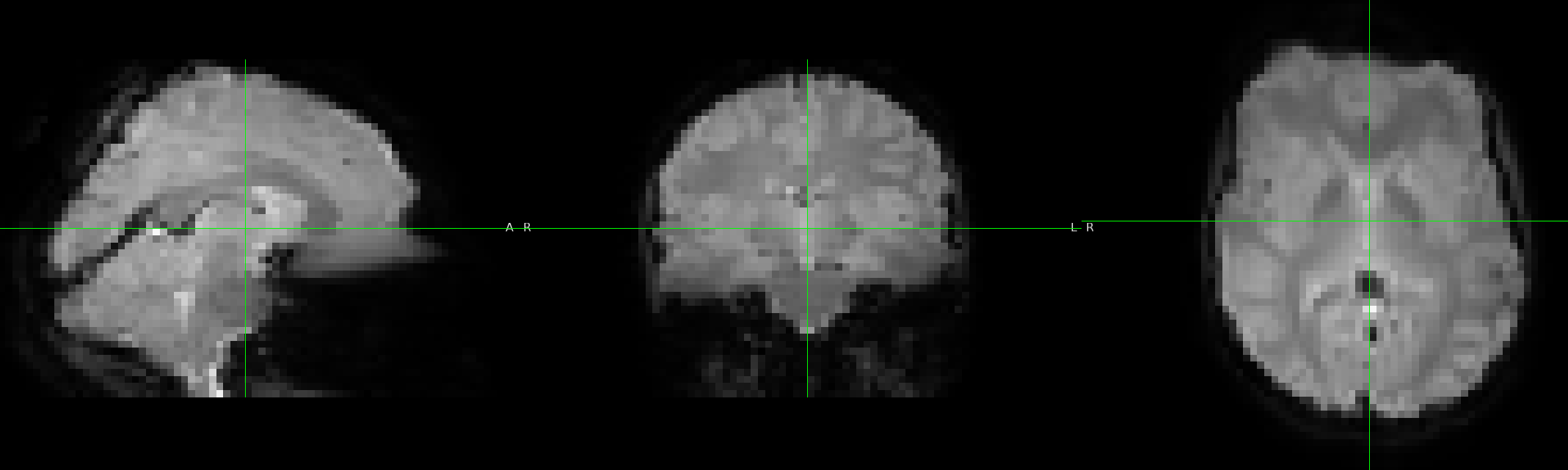
2 fmaps:
no slice timing:
Bold registered to T1 (similar for all cases):
Any suggestions as to what might be the cause? I’m happy to provide any additional information if I forgot anything.
Thanks in advance,
Cheers,
Anthony