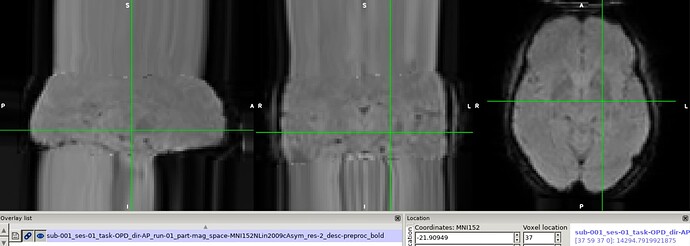
Hi all,
Summary of what happened:
I’ve been processing slab fMRI data with fmriprep. The distortion correction and coregistration (both to T1 and MNI space) look fine but the resulting images have these weirdly protruding voxels in the z-direction:
Anyone have any idea why this happens? Any help greatly appreciated!
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv --contain -B /mnt/projects/OPD/bids/rawdata:/projectFolder -B /mnt/scratch/projects/OPD/bids/derivatives/fmriprep.product.0624-1511:/product -B /mnt/scratch/projects/OPD/bids/derivatives/nobackup/scratch/templateflow2:/templateflow -B /mnt/scratch/projects/OPD/bids/derivatives/fmriprep:/work /mnt/projects/OPD/bids/rawdata/code/singularity/fmriprep-25.1.1.sif --participant-label 001 -w /work --bids-filter-file /projectFolder/filter_file.json --output-spaces MNI152NLin2009cAsym:res-2 --fs-no-reconall --fs-license-file /projectFolder/code/singularity/license.txt /projectFolder /product participant
Version:
25.1.1
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator@1.14.10
e[33m1: [WARN] The Name field of dataset_description.json is present but empty of visible characters. (code: 115 - EMPTY_DATASET_NAME)e[39m
e[36m Please visit Search results for 'EMPTY_DATASET_NAME' - Neurostars for existing conversations about this issue.e[39m
e[33m2: [WARN] The recommended file /README is very small. Please consider expanding it with additional information about the dataset. (code: 213 - README_FILE_SMALL)e[39m
./README.md
e[36m Please visit Search results for 'README_FILE_SMALL' - Neurostars for existing conversations about this issue.e[39m
e[34me[4mSummary:e[24me[39m e[34me[4mAvailable Tasks:e[24me[39m e[34me[4mAvailable Modalities:e[39me[24m
129 Files, 3.34GB OPD MRI
3 - Subjects
1 - Session
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE