Hello fMRIprep gurus!
I have been trying to process HCP data using fMRIprep version 20.2.3
My current BIDS folder structure looks something like this:
sub-100307/anat:
sub-100307_run-01_T1w.nii.gz sub-100307_run-01_T2w.nii.gz
sub-100307/dwi:
sub-100307_acq-dir95lr_dwi.bval sub-100307_acq-dir95rl_dwi.bval sub-100307_acq-dir96lr_dwi.bval sub-100307_acq-dir96rl_dwi.bval sub-100307_acq-dir97lr_dwi.bval sub-100307_acq-dir97rl_dwi.bval
sub-100307_acq-dir95lr_dwi.bvec sub-100307_acq-dir95rl_dwi.bvec sub-100307_acq-dir96lr_dwi.bvec sub-100307_acq-dir96rl_dwi.bvec sub-100307_acq-dir97lr_dwi.bvec sub-100307_acq-dir97rl_dwi.bvec
sub-100307_acq-dir95lr_dwi.json sub-100307_acq-dir95rl_dwi.json sub-100307_acq-dir96lr_dwi.json sub-100307_acq-dir96rl_dwi.json sub-100307_acq-dir97lr_dwi.json sub-100307_acq-dir97rl_dwi.json
sub-100307_acq-dir95lr_dwi.nii.gz sub-100307_acq-dir95rl_dwi.nii.gz sub-100307_acq-dir96lr_dwi.nii.gz sub-100307_acq-dir96rl_dwi.nii.gz sub-100307_acq-dir97lr_dwi.nii.gz sub-100307_acq-dir97rl_dwi.nii.gz
sub-100307_acq-dir95lr_sbref.json sub-100307_acq-dir95rl_sbref.json sub-100307_acq-dir96lr_sbref.json sub-100307_acq-dir96rl_sbref.json sub-100307_acq-dir97lr_sbref.json sub-100307_acq-dir97rl_sbref.json
sub-100307_acq-dir95lr_sbref.nii.gz sub-100307_acq-dir95rl_sbref.nii.gz sub-100307_acq-dir96lr_sbref.nii.gz sub-100307_acq-dir96rl_sbref.nii.gz sub-100307_acq-dir97lr_sbref.nii.gz sub-100307_acq-dir97rl_sbref.nii.gz
sub-100307/fmap:
sub-100307_dir-1_epi.json sub-100307_dir-3_epi.json sub-100307_dir-5_epi.json sub-100307_dir-7_epi.json sub-100307_run-01_magnitude1.nii.gz sub-100307_run-01_phasediff.nii.gz sub-100307_run-02_phasediff.json
sub-100307_dir-1_epi.nii.gz sub-100307_dir-3_epi.nii.gz sub-100307_dir-5_epi.nii.gz sub-100307_dir-7_epi.nii.gz sub-100307_run-01_magnitude2.nii.gz sub-100307_run-02_magnitude1.nii.gz sub-100307_run-02_phasediff.nii.gz
sub-100307_dir-2_epi.json sub-100307_dir-4_epi.json sub-100307_dir-6_epi.json sub-100307_dir-8_epi.json sub-100307_run-01_magnitude.nii.gz sub-100307_run-02_magnitude2.nii.gz
sub-100307_dir-2_epi.nii.gz sub-100307_dir-4_epi.nii.gz sub-100307_dir-6_epi.nii.gz sub-100307_dir-8_epi.nii.gz sub-100307_run-01_phasediff.json sub-100307_run-02_magnitude.nii.gz
sub-100307/func:
sub-100307_task-REST_acq-LR_run-01_bold.nii.gz sub-100307_task-REST_acq-LR_run-02_bold.nii.gz sub-100307_task-REST_acq-RL_run-01_bold.nii.gz sub-100307_task-REST_acq-RL_run-02_bold.nii.gz sub-100307_task-REST_run-01_sbref.nii.gz
sub-100307_task-REST_acq-LR_run-01_sbref.nii.gz sub-100307_task-REST_acq-LR_run-02_sbref.nii.gz sub-100307_task-REST_acq-RL_run-01_sbref.nii.gz sub-100307_task-REST_acq-RL_run-02_sbref.nii.gz sub-100307_task-REST_run-02_sbref.nii.gz
and I have the following .json files on the BIDS parent directory:
task-REST_acq-LR_sbref.json/task-REST_acq-LR_bold.json:
{
"RepetitionTime": 0.72,
"EchoTime": 0.0331,
"EffectiveEchoSpacing": 0.00058,
"MagneticFieldStrength": 3.0,
"Manufacturer": "Siemens",
"ManufacturerModelName": "Skyra",
"PhaseEncodingDirection": "i",
"TaskName": "REST"
}
task-REST_acq-RL_sbref.json/task-REST_acq-LR_sbref.json:
{
"RepetitionTime": 0.72,
"EchoTime": 0.0331,
"EffectiveEchoSpacing": 0.00058,
"MagneticFieldStrength": 3.0,
"Manufacturer": "Siemens",
"ManufacturerModelName": "Skyra",
"PhaseEncodingDirection": "i-",
"TaskName": "REST"
}
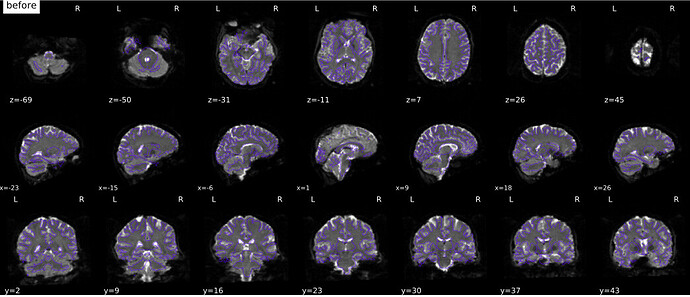
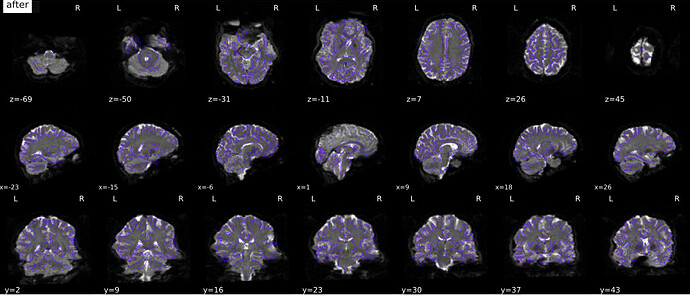
Now, to me the PE directions for these json files appear to be correct. However, the PEB/PEPOLAR SDC seems to be pushing the distortion to the other direction
My question is:
-
Is my json for sbref and bold correct? A LR PE acquisition should be represented by a PED=“i”? Or is that incorrect and it is therefore causing a correction to go in the opposite direction?
-
There are phase and magnitude images in the fmap folder, but I currently have them as intended for the T1w and T2w acquisitions, not the bold acquisitions. Is it possible to use these for SDC?