Hello again,
@AnonymousBoba, thank you for those links.
@oesteban, thank you for sharing you slides on fMRIPrep. Just a few clarification questions regarding the workflow in general, and the use of FreeSurfer.
Our study has two scan sessions, each with its own T1 and subsequent FreeSurfer processed data. We are examining change across time in our specific patient population and expect morphological changes between scan sessions. Therefore, we want to use QC’d FreeSurfer outputs for each session’s T1 in fMRIP prep and separately process fMRI data for each session.
For example,
• Scan session 1: we need to use scan session 1 T1/FreeSurfer for scan session 1 fMRIPrep Processing.
• Scan session 2: we need to use scan session 2 T1/FreeSurfer for scan session 2 fMRIPrep Processing.
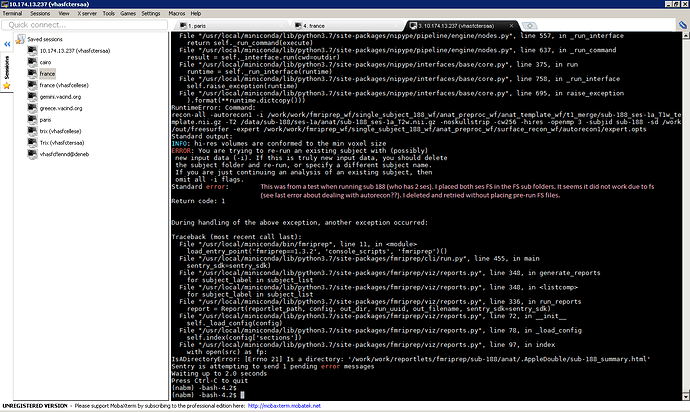
However, our BIDS formatting treats each subject as one subject, with separate scan sessions for each session. Given this file structure, I don’t see a way to split FreeSurfer sub directories in fMRIPrep to use separate T1/FreeSurfer data from each session for processing each session fMRI data.
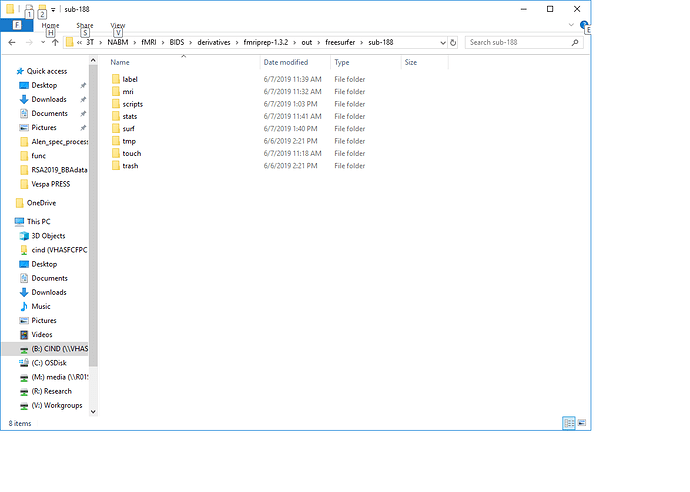
For example, here: > B:\InProcess\3T\NABM\fMRI\BIDS\derivatives\fmriprep-1.3.2\out\freesurfer\sub-123
In the folder structure above, I am unable to place two sessions of pre-run FreeSurfer outputs (i.e. surf, mri, label, etc…) because both session’s folders have the same names (i.e. surf, mri, label, etc…).
So, I tried doing something like this, but it did not work:
B:\InProcess\3T\NABM\fMRI\BIDS\derivatives\fmriprep-1.3.2\out\freesurfer\sub-123\ses-1a
B:\InProcess\3T\NABM\fMRI\BIDS\derivatives\fmriprep-1.3.2\out\freesurfer\sub-123\ses-2a
Is there any way I can ensure each fMRI session uses its session specific pre-run FreeSurfer outputs?
Or is the only alternative to change our naming system in my BIDS script that treats each session as a new subject, for example, something like \sub-1231a and \sub1232a
We would prefer not to have to change our BIDS naming structure in order to use separate T1/FreeSurfer output for each of our sessions. If there is any way to do this without having to take that step, please let us know. I have not been able to find much about this specific issue.
Thank you for the help!