Summary of what happened:
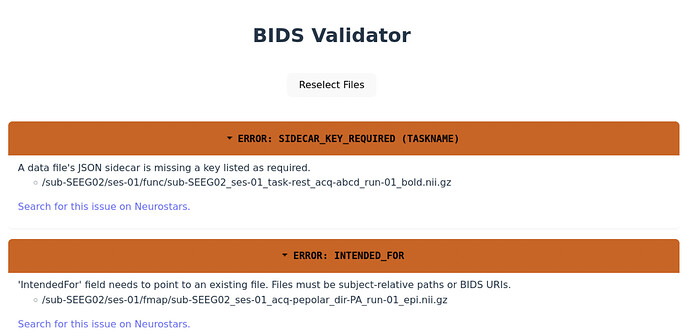
I am encountering an issue in fMRIprep where the Susceptibility Distortion Correction is not being generated. I suspect this is because the field map sequence didn’t convert correctly when I used dcm2bids. Specifically, the ‘IntendedFor’ tags in the .json files seem incorrect. After researching on Neurostars, I found two errors in my field map’s .json file:
- The ‘IntendedFor’ field had a lowercase ‘i’ instead of an uppercase ‘I’.
- The tag contained “bids::sub-SubjectName” as part of the ‘IntendedFor’ field.
Following a suggestion from Neurostars (by Steven), I first removed “bids::sub-SubjectName” from the ‘IntendedFor’ tag in the field map’s .json file and reran fMRIprep. Although fMRIprep ran without errors, the Susceptibility Distortion Correction still did not generate. Next, I changed the lowercase ‘i’ in ‘IntendedFor’ to an uppercase ‘I’, but this caused fMRIprep to throw an error: "bids-validator’ error … returned non-zero exit status 1.
Command used (and if a helper script was used, a link to the helper script or the command generated):
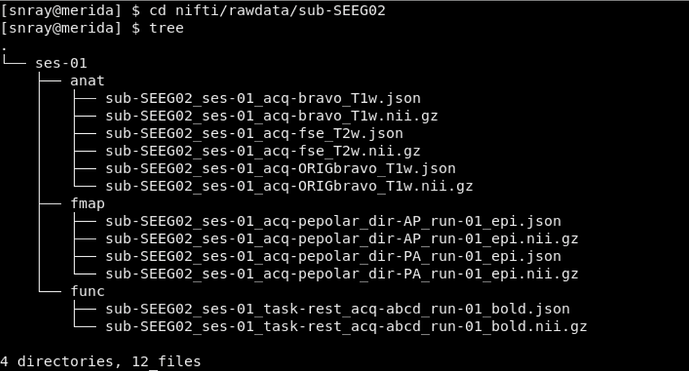
To conver dicom to bids in dcm2bids, following comand I used (Note: SEEG02 is the data folder name):
dcm2bids -d sourcedata/sub-SEEG02/ -p SEEG02 -s 01 -c code/dcm2bids_config.json --auto_extract_entities
For fMRIprep, below comand used:
singularity run --cleanenv \
/data_PATH/software/fmriprep-23.1.4.simg \
/data_PATH/nifti/rawdata
/data_PATH/nifti/derivatives/fmriprep-23.1.4 participant \
--participant-label SEEG02 \
-w /working/snray \
--fs-license-file /home/snray/Documents/freesurfer_license_sr.txt \
--output-spaces T1w MNI152NLin6Asym:res-2 fsaverage5 fsLR:den-32k \
--cifti-output \
--nthreads 6 \
--omp-nthreads 3 \
--verbose
Version:
“Dcm2bidsVersion”: “3.2.0”,
“dcm2niix”: “v1.0.20240202”,
“fmriprep”: 23.1.4
Environment (Docker, Singularity / Apptainer, custom installation):
for dcm2nii, directly in the terminal and for fMRIprep: Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/bin/fmriprep", line 8, in <module>
sys.exit(main())
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/fmriprep/cli/run.py", line 43, in main
parse_args()
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/fmriprep/cli/parser.py", line 812, in parse_args
validate_input_dir(config.environment.exec_env, opts.bids_dir, opts.participant_label)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/fmriprep/utils/bids.py", line 185, in validate_input_dir
subprocess.check_call(['bids-validator', str(bids_dir), '-c', temp.name])
File "/opt/conda/envs/fmriprep/lib/python3.10/subprocess.py", line 369, in check_call
raise CalledProcessError(retcode, cmd)
subprocess.CalledProcessError: Command '['bids-validator', '/data/ucsfocd/data/mri/SEEG_subjects/nifti/rawdata', '-c', '/tmp/tmpkpsinw1e.jsonb']' returned non-zero exit status 1.