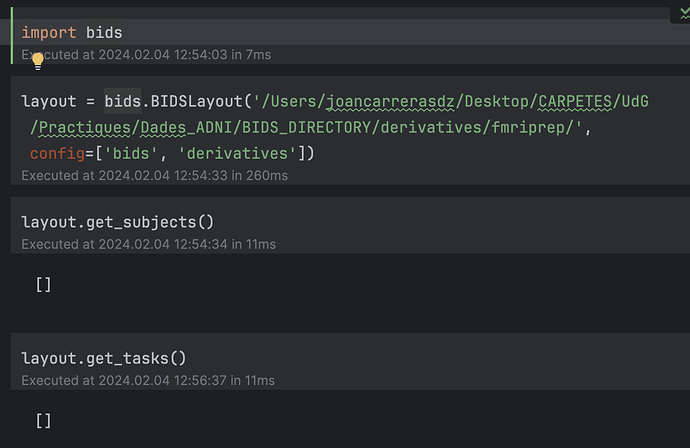
Dear Neurostars team,Functional Neuroimaging Analysis in Python but I am facing some problems when I execute the function layout = bids.BIDSLayout('../data/ds000030/derivatives/fmriprep/', config=['bids','derivatives']). I should have to obtain the list of my subjects once I execute layout.get_subjects()but in order, my result is [ ].
Later in the tutorial, is being said that is necessary to run this function.If we didn’t configure pyBIDS with config=['bids','derivatives'] then the desc keyword would not work!.
Steven
February 5, 2024, 2:02pm
2
Hi @Joan_Carreras_Diaz ,
It would help if we had the BIDS validation report on the BIDS root directory and the tree output of the fmriprep output directory.
Best,
Yes, of course, the tree output of the fmriprep directory is the following one:
/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep
├── dataset_description.json
├── desc-aparcaseg_dseg.tsv
├── desc-aseg_dseg.tsv
├── logs
├── sub-ADNI002S0413
└── sub-ADNI002S0413.html
I don’t think I have any file with the BIDS validation report, as I don’t have any index.html file in the BIDS_DIRECTORY folder:
/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY
├── README
├── conversion_info
├── dataset_description.json
├── derivatives
├── participants.tsv
└── sub-ADNI002S0413
However, I have run bids-validator /Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY and I have obtained the following message:
bids-validator@1.14.0
(node:15960) [DEP0040] DeprecationWarning: The `punycode` module is deprecated. Please use a userland alternative instead.
(Use `node --trace-deprecation ...` to show where the warning was created)
(node:15960) Warning: Closing directory handle on garbage collection
This dataset appears to be BIDS compatible.
Summary: Available Tasks: Available Modalities:
68 Files, 524.32MB MRI
1 - Subject PET
13 - Sessions
If you have any questions, please post on https://neurostars.org/tags/bids.
Thank you,
Steven
February 5, 2024, 3:46pm
4
What about tree run on the fMRIPrep subject output, that is, tree /Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/
The simplification of that would be the following one:
/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/
├── anat
├── figures
├── log
├── ses-M000
├── ses-M006
├── ses-M012
├── ses-M024
├── ses-M036
├── ses-M048
├── ses-M060
├── ses-M072
├── ses-M084
├── ses-M096
├── ses-M108
├── ses-M132
└── ses-M162
If you might prefer the entire one it looks like this:
/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/
├── anat
│ ├── sub-ADNI002S0413_desc-brain_mask.json
│ ├── sub-ADNI002S0413_desc-brain_mask.nii.gz
│ ├── sub-ADNI002S0413_desc-preproc_T1w.json
│ ├── sub-ADNI002S0413_desc-preproc_T1w.nii.gz
│ ├── sub-ADNI002S0413_desc-ribbon_mask.json
│ ├── sub-ADNI002S0413_desc-ribbon_mask.nii.gz
│ ├── sub-ADNI002S0413_dseg.nii.gz
│ ├── sub-ADNI002S0413_from-MNI152NLin2009cAsym_to-T1w_mode-image_xfm.h5
│ ├── sub-ADNI002S0413_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5
│ ├── sub-ADNI002S0413_from-T1w_to-fsnative_mode-image_xfm.txt
│ ├── sub-ADNI002S0413_from-fsnative_to-T1w_mode-image_xfm.txt
│ ├── sub-ADNI002S0413_hemi-L_desc-reg_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-L_midthickness.surf.gii
│ ├── sub-ADNI002S0413_hemi-L_pial.surf.gii
│ ├── sub-ADNI002S0413_hemi-L_space-fsLR_desc-msmsulc_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-L_space-fsLR_desc-reg_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-L_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-L_sulc.shape.gii
│ ├── sub-ADNI002S0413_hemi-L_thickness.shape.gii
│ ├── sub-ADNI002S0413_hemi-L_white.surf.gii
│ ├── sub-ADNI002S0413_hemi-R_desc-reg_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-R_midthickness.surf.gii
│ ├── sub-ADNI002S0413_hemi-R_pial.surf.gii
│ ├── sub-ADNI002S0413_hemi-R_space-fsLR_desc-msmsulc_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-R_space-fsLR_desc-reg_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-R_sphere.surf.gii
│ ├── sub-ADNI002S0413_hemi-R_sulc.shape.gii
│ ├── sub-ADNI002S0413_hemi-R_thickness.shape.gii
│ ├── sub-ADNI002S0413_hemi-R_white.surf.gii
│ ├── sub-ADNI002S0413_label-CSF_probseg.nii.gz
│ ├── sub-ADNI002S0413_label-GM_probseg.nii.gz
│ ├── sub-ADNI002S0413_label-WM_probseg.nii.gz
│ ├── sub-ADNI002S0413_space-MNI152NLin2009cAsym_desc-brain_mask.json
│ ├── sub-ADNI002S0413_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
│ ├── sub-ADNI002S0413_space-MNI152NLin2009cAsym_desc-preproc_T1w.json
│ ├── sub-ADNI002S0413_space-MNI152NLin2009cAsym_desc-preproc_T1w.nii.gz
│ ├── sub-ADNI002S0413_space-MNI152NLin2009cAsym_dseg.nii.gz
│ ├── sub-ADNI002S0413_space-MNI152NLin2009cAsym_label-CSF_probseg.nii.gz
│ ├── sub-ADNI002S0413_space-MNI152NLin2009cAsym_label-GM_probseg.nii.gz
│ └── sub-ADNI002S0413_space-MNI152NLin2009cAsym_label-WM_probseg.nii.gz
├── figures
│ ├── sub-ADNI002S0413_desc-conform_T1w.html
│ ├── sub-ADNI002S0413_desc-reconall_T1w.svg
│ ├── sub-ADNI002S0413_dseg.svg
│ ├── sub-ADNI002S0413_ses-M000_desc-about_T1w.html
│ ├── sub-ADNI002S0413_ses-M000_desc-summary_T1w.html
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-carpetplot_bold.svg
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-compcorvar_bold.svg
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-confoundcorr_bold.svg
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-coreg_bold.svg
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-rois_bold.svg
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-summary_bold.html
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-validation_bold.html
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-carpetplot_bold.svg
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-compcorvar_bold.svg
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-confoundcorr_bold.svg
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-coreg_bold.svg
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-rois_bold.svg
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-summary_bold.html
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-validation_bold.html
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-carpetplot_bold.svg
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-compcorvar_bold.svg
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-confoundcorr_bold.svg
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-coreg_bold.svg
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-rois_bold.svg
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-summary_bold.html
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-validation_bold.html
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-carpetplot_bold.svg
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-compcorvar_bold.svg
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-confoundcorr_bold.svg
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-coreg_bold.svg
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-rois_bold.svg
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-summary_bold.html
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-validation_bold.html
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-carpetplot_bold.svg
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-compcorvar_bold.svg
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-confoundcorr_bold.svg
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-coreg_bold.svg
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-rois_bold.svg
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-summary_bold.html
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-validation_bold.html
│ └── sub-ADNI002S0413_space-MNI152NLin2009cAsym_T1w.svg
├── log
│ ├── 20240117-185138_35bcf3be-da65-485c-a6b4-fb456cf9e865
│ │ ├── crash-20240117-201746-root-anat_merge-b67922a0-fe47-4606-bd80-2ff14f427aa1.txt
│ │ └── fmriprep.toml
│ ├── 20240130-152425_78a06a63-ee48-4bc3-b642-63352eaedc34
│ │ └── fmriprep.toml
│ └── 20240201-191731_98442f86-414d-4962-bfe3-cef64248ea16
│ └── fmriprep.toml
├── ses-M000
│ └── anat
│ └── sub-ADNI002S0413_ses-M000_from-orig_to-T1w_mode-image_xfm.txt
├── ses-M006
│ └── anat
│ └── sub-ADNI002S0413_ses-M006_from-orig_to-T1w_mode-image_xfm.txt
├── ses-M012
│ └── anat
│ └── sub-ADNI002S0413_ses-M012_from-orig_to-T1w_mode-image_xfm.txt
├── ses-M024
│ └── anat
│ └── sub-ADNI002S0413_ses-M024_from-orig_to-T1w_mode-image_xfm.txt
├── ses-M036
│ └── anat
│ └── sub-ADNI002S0413_ses-M036_from-orig_to-T1w_mode-image_xfm.txt
├── ses-M048
│ └── anat
│ └── sub-ADNI002S0413_ses-M048_from-orig_to-T1w_mode-image_xfm.txt
├── ses-M060
│ ├── anat
│ │ └── sub-ADNI002S0413_ses-M060_from-orig_to-T1w_mode-image_xfm.txt
│ └── func
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-confounds_timeseries.json
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-confounds_timeseries.tsv
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-coreg_boldref.json
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-coreg_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-hmc_boldref.json
│ ├── sub-ADNI002S0413_ses-M060_task-rest_desc-hmc_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M060_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
│ ├── sub-ADNI002S0413_ses-M060_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
│ ├── sub-ADNI002S0413_ses-M060_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
│ ├── sub-ADNI002S0413_ses-M060_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
│ ├── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
│ ├── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json
│ └── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
├── ses-M072
│ ├── anat
│ │ └── sub-ADNI002S0413_ses-M072_from-orig_to-T1w_mode-image_xfm.txt
│ └── func
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-confounds_timeseries.json
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-confounds_timeseries.tsv
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-coreg_boldref.json
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-coreg_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-hmc_boldref.json
│ ├── sub-ADNI002S0413_ses-M072_task-rest_desc-hmc_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M072_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
│ ├── sub-ADNI002S0413_ses-M072_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
│ ├── sub-ADNI002S0413_ses-M072_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
│ ├── sub-ADNI002S0413_ses-M072_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
│ ├── sub-ADNI002S0413_ses-M072_task-rest_space-MNI152NLin2009cAsym_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M072_task-rest_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
│ ├── sub-ADNI002S0413_ses-M072_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json
│ └── sub-ADNI002S0413_ses-M072_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
├── ses-M084
│ ├── anat
│ │ └── sub-ADNI002S0413_ses-M084_from-orig_to-T1w_mode-image_xfm.txt
│ └── func
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-confounds_timeseries.json
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-confounds_timeseries.tsv
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-coreg_boldref.json
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-coreg_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-hmc_boldref.json
│ ├── sub-ADNI002S0413_ses-M084_task-rest_desc-hmc_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M084_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
│ ├── sub-ADNI002S0413_ses-M084_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
│ ├── sub-ADNI002S0413_ses-M084_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
│ ├── sub-ADNI002S0413_ses-M084_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
│ ├── sub-ADNI002S0413_ses-M084_task-rest_space-MNI152NLin2009cAsym_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M084_task-rest_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
│ ├── sub-ADNI002S0413_ses-M084_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json
│ └── sub-ADNI002S0413_ses-M084_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
├── ses-M096
│ ├── anat
│ │ └── sub-ADNI002S0413_ses-M096_from-orig_to-T1w_mode-image_xfm.txt
│ └── func
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-confounds_timeseries.json
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-confounds_timeseries.tsv
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-coreg_boldref.json
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-coreg_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-hmc_boldref.json
│ ├── sub-ADNI002S0413_ses-M096_task-rest_desc-hmc_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M096_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
│ ├── sub-ADNI002S0413_ses-M096_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
│ ├── sub-ADNI002S0413_ses-M096_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
│ ├── sub-ADNI002S0413_ses-M096_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
│ ├── sub-ADNI002S0413_ses-M096_task-rest_space-MNI152NLin2009cAsym_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M096_task-rest_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
│ ├── sub-ADNI002S0413_ses-M096_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json
│ └── sub-ADNI002S0413_ses-M096_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
├── ses-M108
│ └── anat
│ └── sub-ADNI002S0413_ses-M108_from-orig_to-T1w_mode-image_xfm.txt
├── ses-M132
│ ├── anat
│ │ └── sub-ADNI002S0413_ses-M132_from-orig_to-T1w_mode-image_xfm.txt
│ └── func
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-confounds_timeseries.json
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-confounds_timeseries.tsv
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-coreg_boldref.json
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-coreg_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-hmc_boldref.json
│ ├── sub-ADNI002S0413_ses-M132_task-rest_desc-hmc_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M132_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
│ ├── sub-ADNI002S0413_ses-M132_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
│ ├── sub-ADNI002S0413_ses-M132_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
│ ├── sub-ADNI002S0413_ses-M132_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
│ ├── sub-ADNI002S0413_ses-M132_task-rest_space-MNI152NLin2009cAsym_boldref.nii.gz
│ ├── sub-ADNI002S0413_ses-M132_task-rest_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
│ ├── sub-ADNI002S0413_ses-M132_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json
│ └── sub-ADNI002S0413_ses-M132_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
└── ses-M162
└── anat
└── sub-ADNI002S0413_ses-M162_from-orig_to-T1w_mode-image_xfm.txt
Hi Joan!
Can you try this:
layout = bids.BIDSLayout('../data/ds000030/derivatives/fmriprep/', derivatives='../data/ds000030/derivatives/fmriprep/', config=['bids','derivatives'])
If I’m correct, what is happening is that BIDSLayout is looking for derivatives under the ./derivatives folder, but since you are setting the main directory to a derivative, it wont find any.
This is pretty counter intuitive, but pybids was not originally conceived to be run on derivatives as first order folders. Obviously, it’s a sensible use case, however.
I don’t really understand your answer, I can’t use this directory as I don’t have these folders. Changing the '../data/ds000030/derivatives/fmriprep/', derivatives='../data/ds000030/derivatives/fmriprep/' for my '/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/' I don’t obtain nothing, but it is because it doesn’t recognize any subject In BIDSLayout.
Oh, sorry, I was just copying your directory in your first question, it should be your directory.
What version of pybids do you have?
My pybids version is 0.16.4
Hmm, I’m not sure what the issue is but can you possibly try the following:
layout = bids.BIDSLayout('/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY', derivatives=True)
1 Like
It has worked! Thank you very much! If there is something else related to this thread I will let you know.
Great! You might try this as well, which is a variation of my original suggestion:
fmriprep_dir = '/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep'
layout = bids.BIDSLayout(fmriprep_dir, derivatives=fmriprep_dir)
I’m guessing the config parameter has changed in how it works since the tutorial was writen.
Yes, it might be this.
When I execute the following commands I don’t notice of any other inconveniences except when I have run the following line that the result is an empty vector:
t1w_func_data = layout.get(datatype='func', desc='preproc', space="T1w")
t1w_func_data
I have noticed that when running the line:
func_data = layout.get(datatype='func', desc='preproc')
func_data
The result is
[<BIDSJSONFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M060/func/sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json'>,
<BIDSImageFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M060/func/sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'>,
<BIDSJSONFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M072/func/sub-ADNI002S0413_ses-M072_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json'>,
<BIDSImageFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M072/func/sub-ADNI002S0413_ses-M072_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'>,
<BIDSJSONFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M084/func/sub-ADNI002S0413_ses-M084_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json'>,
<BIDSImageFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M084/func/sub-ADNI002S0413_ses-M084_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'>,
<BIDSJSONFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M096/func/sub-ADNI002S0413_ses-M096_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json'>,
<BIDSImageFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M096/func/sub-ADNI002S0413_ses-M096_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'>,
<BIDSJSONFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M132/func/sub-ADNI002S0413_ses-M132_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json'>,
<BIDSImageFile filename='/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M132/func/sub-ADNI002S0413_ses-M132_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'>]
As you can see, there is no file named with T1w, however in one of my func folders, there are some files named with T1w:
/Users/joancarrerasdz/Desktop/CARPETES/UdG/Practiques/Dades_ADNI/BIDS_DIRECTORY/derivatives/fmriprep/sub-ADNI002S0413/ses-M060/func
├── sub-ADNI002S0413_ses-M060_task-rest_desc-confounds_timeseries.json
├── sub-ADNI002S0413_ses-M060_task-rest_desc-confounds_timeseries.tsv
├── sub-ADNI002S0413_ses-M060_task-rest_desc-coreg_boldref.json
├── sub-ADNI002S0413_ses-M060_task-rest_desc-coreg_boldref.nii.gz
├── sub-ADNI002S0413_ses-M060_task-rest_desc-hmc_boldref.json
├── sub-ADNI002S0413_ses-M060_task-rest_desc-hmc_boldref.nii.gz
├── sub-ADNI002S0413_ses-M060_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.json
├── sub-ADNI002S0413_ses-M060_task-rest_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt
├── sub-ADNI002S0413_ses-M060_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.json
├── sub-ADNI002S0413_ses-M060_task-rest_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt
├── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_boldref.nii.gz
├── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz
├── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.json
└── sub-ADNI002S0413_ses-M060_task-rest_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
That is expected behavior.
You asked for files with desc-preproc but the T1w files have desc-coreg.
And at least from this list of files, there are no files with space=T1w.