Hi Experts
I have run the AROMA and 2mm resampling grid options with FMRIMPREP, for each of the BOLD MRI, I got the brain mask and two output images:
MNI152NLin2009cAsym_preproc.nii.gz (97x115x97)
MNI152NLin2009cAsym_variant-smoothAROMAnonaggr_preproc.nii.gz (91x109x91)
These two images both have the 2mm voxel size but different dimension as stated above.
These two images lines up perfectly, but with a 2mm atlas I have in the MNI space, even with the same dimension, its voxels could not align with the above 2 images. In each of the axis, they are all of by less than a voxel.
I think this is because of the slight difference in the transformation matrices.
Comparing the non-aggressive AROMA output which has the same voxel dimension with my atlas (91x109x91),
the MNI152NLin2009cAsym_variant-smoothAROMAnonaggr_preproc.nii.gz has the following transformation matrix:
2 0 0 -90.5
0 2 0 -126.5
0 0 2 -72.5
0 0 0 1
and the transformation of my atlas:
-2 -0 -0 90
-0 2 -0 -126
0 0 2 -72
0 0 0 1
Could anyone please suggest on how I could solve the problem?
Thank you very much!
Could you share the particular MNI atlas that you experience problems with?
Also providing a single example volume from each of the FMRIPREP outputs you mentioned would also help to diagnose the problem.
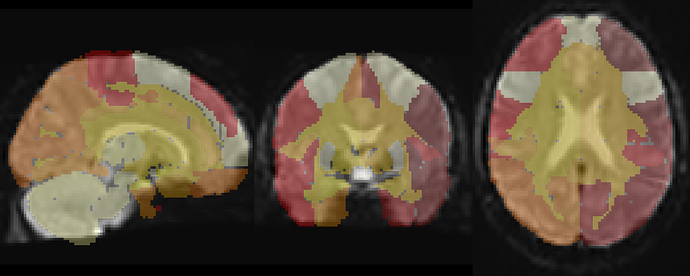
The two output images after using fslmaths -Tmean (the transformation matrices remained the same as the 4D volumes) and the atlas.
Could I email it to you because I can’t seemed to be able to upload .nii.gz here?
Thank you!
Sure. chrisgor@stanford.edu
Our atlas is in RAS orientation while the original MNI files are in LAS. Visualization tools should deal with this issue.
Otherwise, you could convert either the AROMA results or your atlas to the alternative orientation. Nibabel is really handy for that.
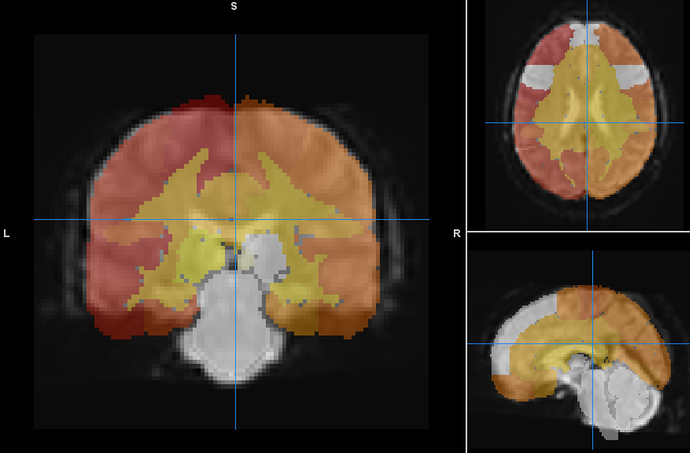
Thanks for sending the example data. I overlaid the atlas you provided on top of the MNI152NLin2009cAsym_preproc file using Mango viewer
This looks like a pretty good alignment to me. Could you describe how did you assess the alignment (which software did you use?
I used fsleyes.

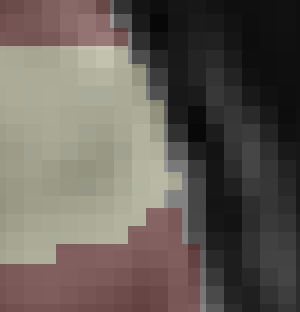
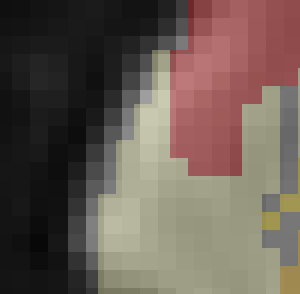
So I guess it was related to the viewer?
Thank you!
Hi @oesteban
Is this the reason why when I tried to fslmaths to add the output and the atlas,
it showed:
“WARNING:: Inconsistent orientations for individual images in pipeline!
Will use voxel-based orientation which is probably incorrect - PLEASE CHECK!”?
Thank you!
This is so subtle I’m not sure it is really missaligned.
One more thing to check is to see how well your atlas overlaps with the ICBM 2009c Nonlinear Asymmetric template (which defines the particular variant of the MNI space used in FMRIPREP). You can find this template at http://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009
Yes, I’d say so. I don’t think fslmaths takes care of the orientation and thus just shows a WARNING.
What about the slight difference of 0.5 in the transformation matrix?
Thank you!
What you are seeing in fsleyes is a consequence of a particular way this software is dealing with resampling (or rather lack of it). As a more general point one should not expect to achieve alignment better than the resolution of the image (in your case the difference is 0.5mm and the image resolution is 2mm).
Why your atlas has (0,0,0) coordinate in an 0.5mm different location than the template we used that I don’t know (it’s probably a consequence of how the template was created). You can try to change the header of your atlas to add the 0.5mm and see if this leads to better alignment, but in general, the 0.5mm difference should be negligible considering the resolution of the image.
That offset is happening because the coordinate system of the original MNI atlas (LAS) is not exactly centered at the center of the imaged volume.