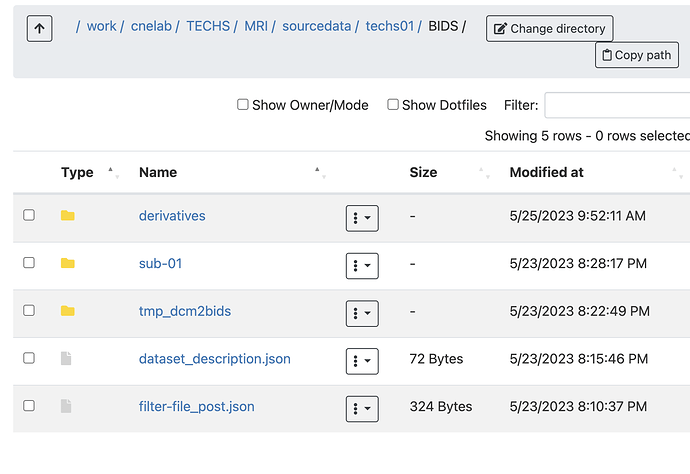
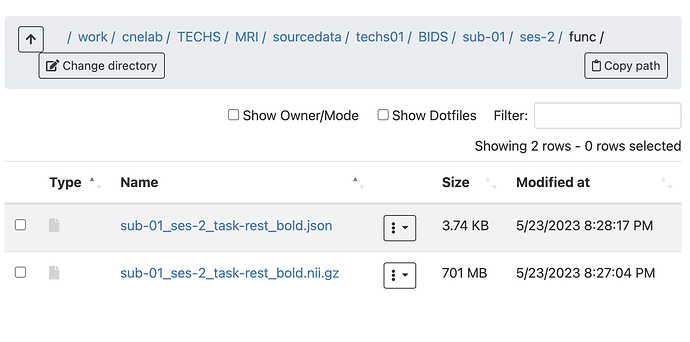
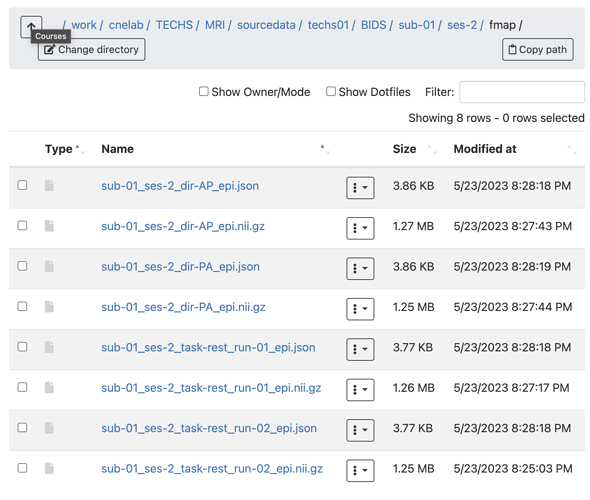
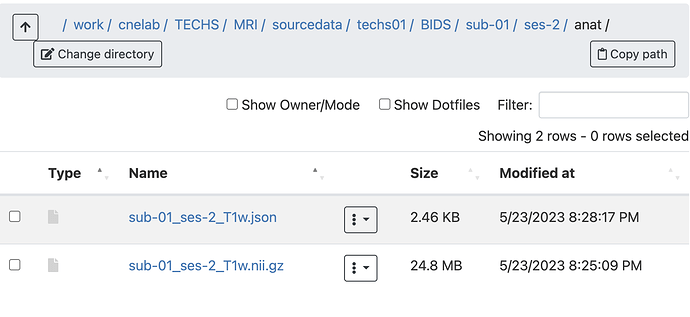
Summary of what happened:
I am getting an error when running fmriprep. “Node Name: fmriprep_23_0_wf.single_subject_01_wf.syn_preprocessing_auto_00001.epi_reference_wf.validate_nii”
everything else seemed to run okay and the output looks accurate from the html file.
Command used (and if a helper script was used, a link to the helper script or the command generated):
/opt/conda/bin/fmriprep /data /data//derivatives/fmriprep participant --participant-label 01 -w /work/ -vv --omp-nthreads 8 --nthreads 12 --mem_mb 30000 --output-spaces MNI152NLin2009cAsym:res-2 anat fsnative fsaverage5 --use-aroma --fs-subjects-dir /fsdir --bids-filter-file /data/filter-file_post.json
Version:
fmriprep 23.0.2
Environment (Docker, Singularity, custom installation):
singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS validated
Relevant log outputs (up to 20 lines):
Errors
Node Name: fmriprep_23_0_wf.single_subject_01_wf.syn_preprocessing_auto_00001.epi_reference_wf.validate_nii
File: /data/derivatives/fmriprep/sub-01/log/20230523-203835_cf0aba38-5e2b-477c-92cd-70bb4461a679/crash-20230523-204005-tinney.e-validate_nii-0060c27e-1190-4770-9799-16eaa469988c.txt
Working Directory: /work/fmriprep_23_0_wf/single_subject_01_wf/syn_preprocessing_auto_00001/epi_reference_wf/validate_nii
Inputs:
in_file:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 292, in _send_procs_to_workers
num_subnodes = self.procs[jobid].num_subnodes()
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 1309, in num_subnodes
self._check_iterfield()
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 1332, in _check_iterfield
raise ValueError(
ValueError: Input in_file was not set but it is listed in iterfields.
When creating this crashfile, the results file corresponding
to the node could not be found.
Node Name: fmriprep_23_0_wf.single_subject_01_wf.syn_preprocessing_auto_00002.epi_reference_wf.validate_nii
File: /data/derivatives/fmriprep/sub-01/log/20230523-203835_cf0aba38-5e2b-477c-92cd-70bb4461a679/crash-20230523-204005-tinney.e-validate_nii-27b4e1de-c601-420b-b675-bda872d3f259.txt
Working Directory: /work/fmriprep_23_0_wf/single_subject_01_wf/syn_preprocessing_auto_00002/epi_reference_wf/validate_nii
Inputs:
in_file:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 292, in _send_procs_to_workers
num_subnodes = self.procs[jobid].num_subnodes()
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 1309, in num_subnodes
self._check_iterfield()
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 1332, in _check_iterfield
raise ValueError(
ValueError: Input in_file was not set but it is listed in iterfields.
When creating this crashfile, the results file corresponding
to the node could not be found.