Summary of what happened:
Hi all,
I am using fmriprep to preprocess resting state and task functional images, but I cannot seem to get the field mapping images to apply for distortion correction.
We are using a short 2 volume reverse-direction “TOPUP” type scan to correct the full-length images.
The fmap images are in proper bids format and bound to the fmriprep /data/input variable, and the “IntendedFor” and “RawSources” fields are added to the associated .json file but when I run the fmriprep workflow, this shows in the log and the images are visually not field-map corrected:
nipype.interface INFO:
No "fmap" images found for sub-P124
Any recommendations or solutions?
Command used (and if a helper script was used, a link to the helper script or the command generated):
inputdir=/project/oathes_analysis2/R61/bids
outputdir=/project/oathes_analysis2/R61/fmriprep/prerun_output/$1
singularity run --cleanenv \
--no-home \
-B ${jobTmpDir}:/tmp \
-B "/project/oathes_analysis2/templateflow:/templateflow" \
-B "/appl/freesurfer-7.1.1:/freesurfer" \
-B ${inputdir}:/data/input \
-B ${outputdir}:/data/output \
-B /project/oathes_analysis2/R61/fmriprep/prerun_output/$1:/fssubdir \
-B /project/oathes_analysis2/individual_projects/jess/R61_tmsfmri_fmriprep_test:/basepath \
/project/oathes_analysis2/singularity_containers/fmriprep-23.2.3.simg \
/data/input /data/output participant --skull-strip-template OASIS30ANTs --fs-license-file /freesurfer/license.txt \
--bids-filter-file /basepath/bids_filter_file_tmsfmri.json \
--fs-subjects-dir /fssubdir/sourcedata/freesurfer \
--output-spaces fsaverage T1w fsnative fsLR MNI152NLin6Asym:res-2 --cifti-output 91k \
--bold2t1w-dof 6 \
--dvars-spike-threshold 1.5 \
--fd-spike-threshold 0.5 \
--ignore slicetiming \
--notrack --nthreads 16 --omp-nthreads 15 --work-dir ${SINGULARITYENV_TMPDIR} --verbose --participant-label $1
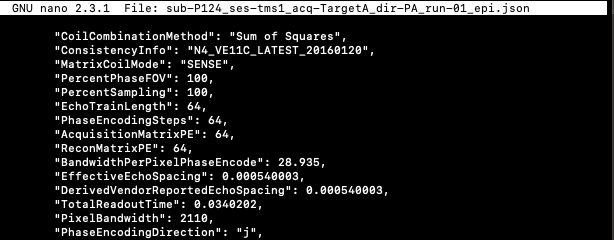
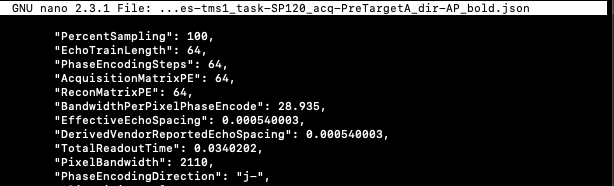
IntendedFor & RawSources example:
"IntendedFor": ["ses-tms1/func/sub-P124_ses-tms1_task-rest_acq-PreTargetA_dir-AP_bold.nii.gz", "ses-tms1/func/sub-P124_ses-tms1_task-SP120_acq-PostTargetA_dir-AP_bold.nii.gz","ses-tms1/func/sub-P124_ses-tms1_task-SP120_acq-PreTargetA_dir-AP_bold.nii.gz", "ses-tms1/func/sub-P124_ses-tms1_task-iTBS_acq-TargetA_dir-AP_bold.nii.gz"],
"RawSources": ["data/input/sub-P124/ses-tms1/fmap/sub-P124_ses-tms1_acq-TargetA_dir-PA_run-01_epi.nii.gz"]
Version:
fmriprep-23.2.3
(I know that this is not the most up-to-date version, but we are using this version for baseline processing, and would prefer to keep it consistent if possible for now)
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity/Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator@1.14.6
1: [WARN] The recommended file /README is missing. See Section 03 (Modality agnostic files) of the BIDS specification. (code: 101 - README_FILE_MISSING)
2: [WARN] The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered because there are no authors, which will make DOI registration from dataset metadata impossible. (code: 113 - NO_AUTHORS)
├── ses-tms1
│ ├── anat
│ ├── fmap
│ │ ├── sub-P124_ses-tms1_acq-TargetA_dir-PA_run-01_epi.json
│ │ ├── sub-P124_ses-tms1_acq-TargetA_dir-PA_run-01_epi.nii.gz
│ │ ├── sub-P124_ses-tms1_acq-TargetB_dir-PA_run-01_epi.json
│ │ ├── sub-P124_ses-tms1_acq-TargetB_dir-PA_run-01_epi.nii.gz
│ │ ├── sub-P124_ses-tms1_acq-Vertex_dir-PA_run-01_epi.json
│ │ └── sub-P124_ses-tms1_acq-Vertex_dir-PA_run-01_epi.nii.gz
│ └── func
│ ├── sub-P124_ses-tms1_task-iTBS_acq-TargetA_dir-AP_bold.json
│ ├── sub-P124_ses-tms1_task-iTBS_acq-TargetA_dir-AP_bold.nii.gz
│ ├── sub-P124_ses-tms1_task-iTBS_acq-TargetB_dir-AP_bold.json
│ ├── sub-P124_ses-tms1_task-iTBS_acq-TargetB_dir-AP_bold.nii.gz
│ ├── sub-P124_ses-tms1_task-rest_acq-PreTargetA_dir-AP_bold.json
│ ├── sub-P124_ses-tms1_task-rest_acq-PreTargetA_dir-AP_bold.nii.gz
│ ├── sub-P124_ses-tms1_task-SP120_acq-PostTargetA_dir-AP_bold.json
│ ├── sub-P124_ses-tms1_task-SP120_acq-PostTargetA_dir-AP_bold.nii.gz
│ ├── sub-P124_ses-tms1_task-SP120_acq-PostTargetB_dir-AP_bold.json
│ ├── sub-P124_ses-tms1_task-SP120_acq-PostTargetB_dir-AP_bold.nii.gz
│ ├── sub-P124_ses-tms1_task-SP120_acq-PreTargetA_dir-AP_bold.json
│ ├── sub-P124_ses-tms1_task-SP120_acq-PreTargetA_dir-AP_bold.nii.gz
│ ├── sub-P124_ses-tms1_task-SP120_acq-PreTargetB_dir-AP_bold.json
│ ├── sub-P124_ses-tms1_task-SP120_acq-PreTargetB_dir-AP_bold.nii.gz
│ ├── sub-P124_ses-tms1_task-SP120_acq-Vertex_dir-AP_bold.json
│ └── sub-P124_ses-tms1_task-SP120_acq-Vertex_dir-AP_bold.nii.gz
Relevant log outputs (up to 20 lines):
nipype.interface INFO:
No "fmap" images found for sub-P124