Hi, Dear all,
i have validated my BIDs data on the validator website, and there is not error messages.
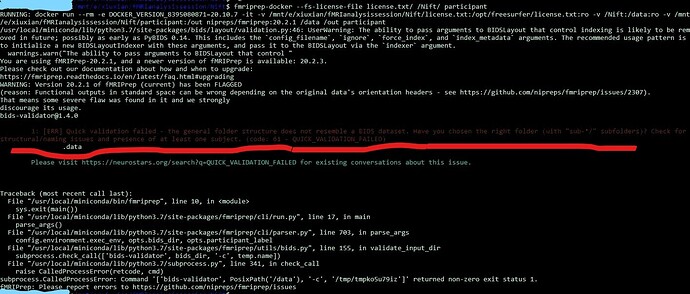
so i start fmriprep, as the followed image. the error comes out like this:
1: [ERR] Quick validation failed - the general folder structure does not resemble a BIDS dataset. Have you chosen the right folder (with “sub-*/” subfolders)? Check for structural/naming issues and presence of at least one subject. (code: 61 - QUICK_VALIDATION_FAILED)
have any ideas to solve this problem?
thank you for your help.
greetings,
chen
There should be three positional (that is, non-named) arguments. I only see two (/Nifti/ and participant) it looks like you need to specify the output directory after /Nifti/. Also, can you show us the output of ls /Nifti/ so we can confirm it looks BIDS-compliant? Finally, you might want to update to 20.2.3 before running again, since 20.2.1 has been flagged. Hope this helps.
Best,
Steven
Hi, Steven,
Thank you for the reply.
I have solved the problem I forgot to put license.txt file in a folder separated from Nifti folder.
And BTW, do you think 20.2.1 and 20.2.3 function different?
since i make it run with 20.2.1, and there are output files.
but one thing confused me is: at the end of the processure after the long time running,
an error comes out like this:
File “/usr/local/miniconda/lib/python3.7/multiprocessing/forkserver.py”, line 261 in main
File “”, line 1 in
ERRO[40676] error waiting for container: invalid character ‘u’ looking for beginning of value
fMRIPrep: Please report errors to Issues · nipreps/fmriprep · GitHub
is it finally finished. Should i use the results from output?
Thank you for reply.
Greetings
Chen
20.2.3 fixes some problems in 20.2.1 for sampling to standard spaces. Unless you’ve already processed all of your data and are happy with the results, I’d suggest updating.
You’ll know it’s finished when you see a subject-specific HTML file in the output directory. You can open that and look at the results and see if you are happy with them. It is hard to diagnose that error without knowing the full command you submitted and the full traceback of the error.
Best,
Steven
Thank you! I make it works!
Greetings
chan