Dear experts,
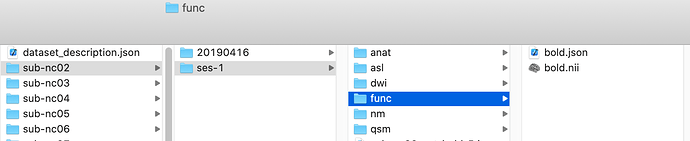
I did a test run on sub-nc02 using fmriPrep using docker on our server. The input T1w and bold image are the same with bids format. The pipeline seems to work fine on T1w process, but no bold analysis is done. I attached the input data structure and output folder.
[environment]
cpu_count = 144
exec_env = “docker”
free_mem = 30.6
overcommit_policy = “heuristic”
overcommit_limit = “50%”
nipype_version = “1.6.1”
templateflow_version = “0.7.2”
version = “21.0.1”
The code I used to run the pipeline is:
docker run -d --rm -v /projects/transimg/Projects/raw_nifti:/data:ro -v /projects/transimg/Projects/output:/out -v /projects/transimg/freesurfer/license.txt:/license nipreps/fmriprep:latest --skip_bids_validation --fs-license-file /license /data /out participant --participant_label sub-nc02 --write-graph
I cannot attach any error logs here, but I will paste one as an example:
Node: fmriprep_wf.single_subject_nc02_wf.func_preproc_.func_derivatives_wf.ds_bold_std
Working directory: /tmp/work/fmriprep_wf/single_subject_nc02_wf/func_preproc_/func_derivatives_wf/_in_tuple_MNI152NLin2009cAsym.resnative/ds_bold_std
Node inputs:
acquisition =
atlas =
base_directory = /out
ceagent =
check_hdr = True
cohort =
compress = [True]
data_dtype =
datatype =
density =
desc = preproc
direction =
dismiss_entities =
echo =
extension =
fmap =
fmapid =
from =
hemi =
in_file = [’/tmp/work/fmriprep_wf/single_subject_nc02_wf/func_preproc_/bold_std_trans_wf/_std_target_MNI152NLin2009cAsym.resnative/merge/vol0000_xform-00000_clipped_merged.nii.gz’]
label =
meta_dict =
modality =
mode =
model =
proc =
reconstruction =
recording =
resolution = native
roi =
run =
scans =
session =
source_file = [’/data/sub-nc02/ses-1/func/bold.nii’]
space = MNI152NLin2009cAsym
subject =
subset =
suffix =
task =
to =
Traceback (most recent call last):
File “/opt/conda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py”, line 344, in _send_procs_to_workers
self.procs[jobid].run(updatehash=updatehash)
File “/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py”, line 516, in run
result = self._run_interface(execute=True)
File “/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py”, line 635, in _run_interface
return self._run_command(execute)
File “/opt/conda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py”, line 741, in _run_command
result = self._interface.run(cwd=outdir)
File “/opt/conda/lib/python3.8/site-packages/nipype/interfaces/base/core.py”, line 428, in run
runtime = self._run_interface(runtime)
File “/opt/conda/lib/python3.8/site-packages/niworkflows/interfaces/bids.py”, line 533, in _run_interface
in_entities = [
File “/opt/conda/lib/python3.8/site-packages/niworkflows/interfaces/bids.py”, line 534, in
parse_file_entities(str(relative_to_root(source_file)))
File “/opt/conda/lib/python3.8/site-packages/niworkflows/utils/bids.py”, line 392, in relative_to_root
raise ValueError(
ValueError: Could not determine the BIDS root of </data/sub-nc02/ses-1/func/bold.nii>. Only files under a subject directory are currently supported.
Thank you in advance for your help!
Best,
Yuchuan