I used fmriprep to pre-process my dataset but I noticed after running GLM analysis on the data that there was activation outside of the anatomical underlay. I found that the bold data did not align with the anatomical. Any idea what is causing this issue or any suggestions for options to run in fmriprep to better align the anatomical and functional data? I have attached images below of the misalignment below. The underlay image is the MNI152NLin2009cAsym_boldref.nii.gz from one of my runs and the overlay is the MNI152NLin2009cAsym_desc-brain_mask.nii.gz from the anatomical data.
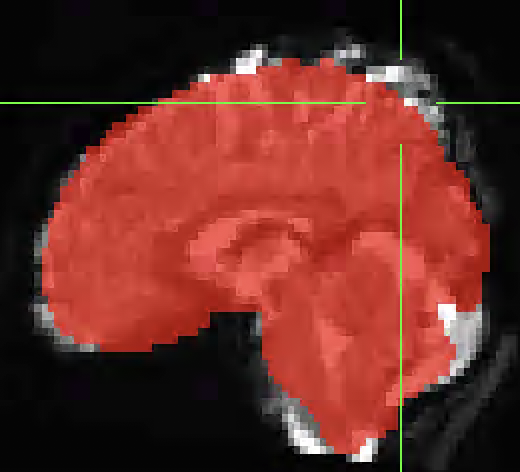
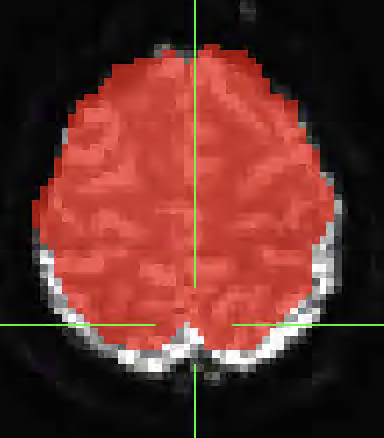
How does the alignment between the T1w and BOLD look in the corresponding report? could you send us the corresponding report (incl. the figures folder)?
That said (reports are the first line of defense when checking the quality of preprocessing), did you run susceptibility distortion correction? If not, that is likely the cause of those misalignments. To better check this, you might want to overlay the white matter mask of the template instead of the whole-brain mask.
The report looks fine from what I can tell. I cannot upload the file here, but I will open an issue on github and upload the report there. The susceptibility distortion was run.
Shown below, the overlay of the anatomical WM mask on the registered bold data does not match up.
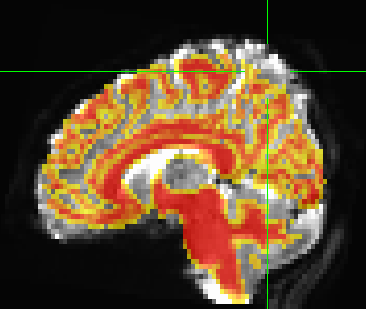
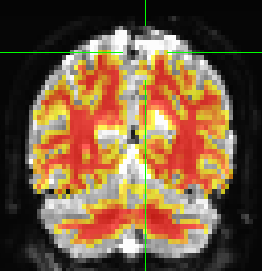
Looks to me that the ventricle and the corpus callosum are pretty tightly matched. Superior gyri are not horrible either. You have, however, mismatched the frontal pole and the parietal - I’d say that most of the problem is susceptibility distortions.
In other words, because your BOLD-EPI data are still distorted (due to susceptibility inhomogeneities), they will never align correctly with the corresponding T1w anatomical image (i.e., distortion-free).
Adding one more nonlinear transformation step (from the T1w to the template) just exacerbates the issue.
The only solution to this problem, and I still need to check on your reports, is to actually run susceptibility distortion. Preferably using fieldmaps, but you can also give SDC-SyN a try.
For more details, please check https://fmriprep.org/en/stable/sdc.html
1 Like
After looking at your reports I don’t think this is a problem. Your BOLD data have a noisy area around the commissure between parietal lobes. But the BOLD-T1w looks okay to me (not impressive, TBH, most of the time I feel the alignment is better than in your case) and the T1w-MNI alignment is also okay.
The problem seems exacerbated by a not-great distortion correction. I think you could expect some minimal improvements with better versions of the SDC map estimation workflow (see the https://github.com/nipreps/sdcflows project), but other than that, most of the signal we see in your original neurostars post is basically noise. I don’t think it is to worry, honestly.
Okay thanks for your help! We are going to take your advice and not worry about it. We are only really looking at the thalamus anyway but wanted to see why the misalignment was occurring and if we could fix it.



