Summary of what happened:
I’m working with glioma patient data, on which FreeSurfer normally doesn’t work very well; but thanks to recon-all-clinical, using FreeSurfer on our data is now possible.
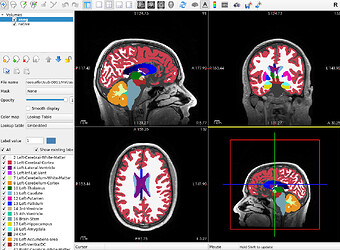
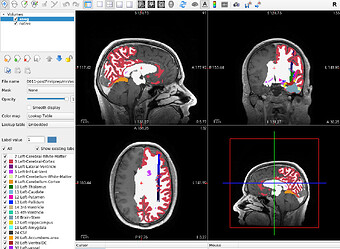
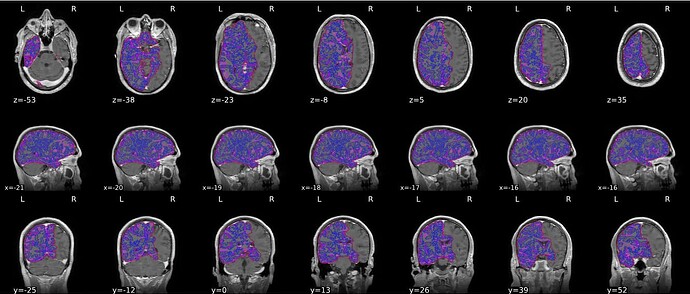
However, providing the outputs of recon-all-clinical to fMRIPrep yields some strange results: fMRIPrep seems to redo/alter FreeSurfer’s aseg.mgz, ignoring a large chunk of brain, and the brain mask used by fMRIPrep is equally bad-- please see the screenshots included below.
Is this a known issue with the relatively new recon-all-clinical, and is there any way to get this to work?
Any help would be greatly appreciated!
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv -B $BIDSDIR -B $OUTDIR -B $WORKDIR -B $FS_LICENSE -B $FSDIR -B $BIDS_CONFIG_FPREP $FPREP_IMG \
$BIDSDIR $OUTDIR participant \
--participant-label $SUBJ_ID \
--use-aroma \
--output-spaces func T1w fsnative fsaverage5 MNI152NLin6Asym:res-2 \
--skip-bids-validation \
--fs-license-file $FS_LICENSE \
--fs-subjects-dir $FSDIR \
--bids-filter-file $BIDS_CONFIG_FPREP \
--ignore fieldmaps slicetiming \
--use-syn-sdc \
--stop-on-first-crash \
--omp-nthreads 8 \
--nthreads 16 \
--mem-mb 32000 \
--work-dir $WORKDIR
Version:
22.0.2
Environment (Docker, Singularity, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
Yes
Relevant log outputs (up to 20 lines):
N/A; fMRIPrep finishes without errors
Screenshots / relevant information:
Initial aseg.mgz after running recon-all-clinical
aseg.mgz after running fMRIPrep
fMRIPrep T1w brain mask & tissue segmentation
Best!
Lucas