Thank you so much,
I have not tried the new version yet since I think my command has a problem,
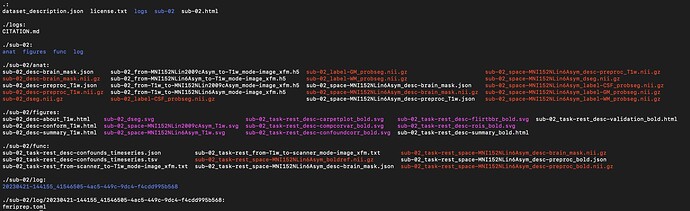
here is the content of JSON files:
bold:
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 127.764,
"Manufacturer": "Philips",
"ManufacturersModelName": "Ingenia",
"InstitutionName": "CHUS FLEURIMONT Philips 3t",
"InstitutionalDepartmentName": "IRM",
"DeviceSerialNumber": "71081",
"StationName": "PHILIPS-TQJ73JP",
"BodyPartExamined": "BRAIN",
"PatientPosition": "HFS",
"SoftwareVersions": "5.4.1\\5.4.1.1",
"MRAcquisitionType": "2D",
"SeriesDescription": "fMRI MB4 3mm",
"ProtocolName": "WIP fMRI MB4 3mm",
"ScanningSequence": "GR",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"FFE",
"M",
"FFE"
],
"SeriesNumber": 301,
"PhaseEncodingDirection":"j",
"AcquisitionTime": "11:31:4.910000",
"AcquisitionNumber": 3,
"PhilipsRWVSlope": 0.867155,
"PhilipsRWVIntercept": 0,
"PhilipsRescaleSlope": 0.867155,
"PhilipsRescaleIntercept": 0,
"PhilipsScaleSlope": 0.0240969,
"UsePhilipsFloatNotDisplayScaling": 1,
"SliceThickness": 3,
"SpacingBetweenSlices": 3,
"SAR": 0.045679,
"EchoTime": 0.03,
"RepetitionTime": 1.075,
"SliceTiming":[0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000,0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000,0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000,0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000],
"MTState": false,
"FlipAngle": 55,
"CoilString": "MULTI COIL",
"PercentPhaseFOV": 100,
"PercentSampling": 100,
"EchoTrainLength": 67,
"PhaseEncodingSteps": 80,
"AcquisitionMatrixPE": 80,
"ReconMatrixPE": 80,
"ParallelReductionFactorInPlane": 1.2,
"ParallelAcquisitionTechnique": "SENSE",
"WaterFatShift": 17.7943,
"EffectiveEchoSpacing": 0.000510896,
"TotalReadoutTime": 0.0403608,
"PixelBandwidth": 2241,
"PhaseEncodingAxis": "j",
"ImageOrientationPatientDICOM": [
0.995206,
-0.0132422,
0.0969009,
0.0398551,
0.959703,
-0.278176
],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20211220",
"Dcm2bidsVersion": "2.1.6"
}
and epi:
{>
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 127.764,
"Manufacturer": "Philips",
"ManufacturersModelName": "Ingenia",
"InstitutionName": "CHUS FLEURIMONT Philips 3t",
"InstitutionalDepartmentName": "IRM",
"DeviceSerialNumber": "71081",
"StationName": "PHILIPS-TQJ73JP",
"BodyPartExamined": "BRAIN",
"PatientPosition": "HFS",
"SoftwareVersions": "5.4.1\\5.4.1.1",
"MRAcquisitionType": "2D",
"SeriesDescription": "rev fMRI MB4 3mm",
"ProtocolName": "WIP rev fMRI MB4 3mm",
"ScanningSequence": "GR",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"FFE",
"M",
"FFE"
],
"SeriesNumber": 401,
"AcquisitionTime": "11:41:43.190000",
"AcquisitionNumber": 4,
"PhilipsRWVSlope": 0.793895,
"PhilipsRWVIntercept": 0,
"PhilipsRescaleSlope": 0.793895,
"PhilipsRescaleIntercept": 0,
"PhilipsScaleSlope": 0.0244135,
"UsePhilipsFloatNotDisplayScaling": 1,
"SliceThickness": 3,
"SpacingBetweenSlices": 3,
"SAR": 0.0452711,
"EchoTime": 0.030001,
"RepetitionTime": 1.075,
"SliceTiming":[0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000,0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000,0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000,0,0.0447916500000000,0.0895833000000000,0.246354075000000,0.201562425000000,0.156770775000000,0.0223958250000000,0.0671874750000000,0.111979125000000,0.223958250000000,0.179166600000000,0.134374950000000],
"MTState": false,
"FlipAngle": 55,
"CoilString": "MULTI COIL",
"PercentPhaseFOV": 100,
"PercentSampling": 100,
"EchoTrainLength": 67,
"PhaseEncodingSteps": 80,
"AcquisitionMatrixPE": 80,
" PhaseEncodingDirection":"j-",
"ReconMatrixPE": 80,
"ParallelReductionFactorInPlane": 1.2,
"ParallelAcquisitionTechnique": "SENSE",
"WaterFatShift": 17.6939,
"EffectiveEchoSpacing": 0.000508013,
"TotalReadoutTime": 0.040133,
"PixelBandwidth": 2318,
"PhaseEncodingAxis": "j",
"ImageOrientationPatientDICOM": [
0.995206,
-0.0132422,
0.0969009,
0.0398551,
0.959703,
-0.278176
],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20211220",
"Dcm2bidsVersion": "2.1.6",
"IntendedFor": "func/sub-02_task-rest_bold.nii.gz"
}
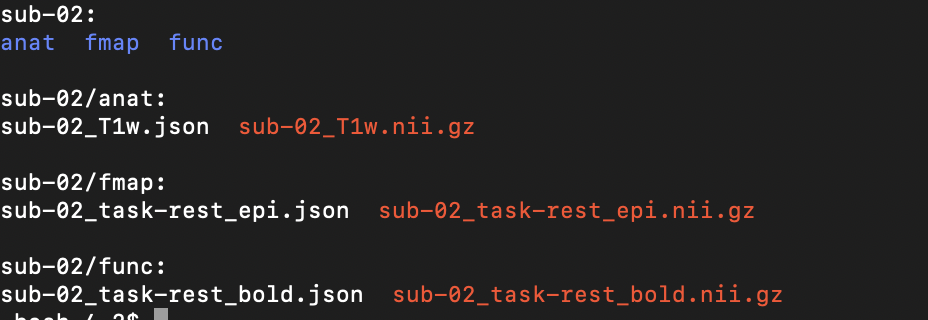
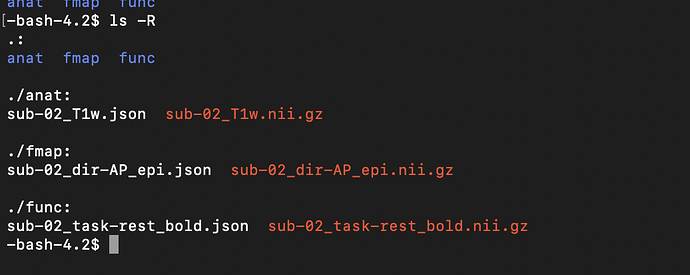
and the output tree: