Summary of what happened:
I processed task fMRI data from a sample of 170 participants who were all scanned on the same scanner with the same sequences. Specifically, we had 6 runs of a task and acquired 4 top-up scans with the opposite phase encoding direction before each block. For two participants, I get an error that sdecreg_report failed for one or two blocks. I cannot find any difference between these blocks or participants and the rest of the sample.
It’s seems like a small issue, because I could just exclude the two participants. However, I’m worried that this may indicate some more fundamental issues with the data.
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep-docker \
${bids_folder} \
${out_folder} \
participant \
--participant-label sub-003 \
--image nipreps/fmriprep:23.2.1 \
--fs-license-file ${license_file} \
--cifti-output \
--nthreads 5
Version:
I processed all participants with version 23.2.1, but also tried 24.1.1 with the problematic participants. The error was the same in both versions.
Environment (Docker, Singularity / Apptainer, custom installation):
Docker using the fmriprep-docker wrapper
Data formatted according to a validatable standard? Please provide the output of the validator:
[WARN] Not all subjects/sessions/runs have the same scanning parameters
[WARN] The onset of the last event is after the total duration of the corresponding scan. This design is suspiciously long.
The scan duration is not the same for all scans, because it depended on the response duration of the participants.
Relevant log outputs (up to 20 lines):
File: /out/sub-003/log/20250219-152253_2d164a42-c1aa-430e-bcc0-0c1641d54df6/crash-20250219-181842-root-sdecreg_report-ec36e71e-0822-4c84-908f-6d9d44956f13.txt
Working Directory: /tmp/work/fmriprep_23_2_wf/sub_003_wf/bold_task_raven_run_1_wf/bold_fit_wf/func_fit_reports_wf/sdecreg_report
Inputs:
apply_mask: False
fieldmap:
mask:
max_alpha: 0.7
moving:
moving_label: Fieldmap reference
out_report: report.svg
reference:
reference_label: BOLD reference
show: both
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node sdecreg_report.
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._post_run_hook(runtime)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/mixins/reporting.py", line 50, in _post_run_hook
self._generate_report()
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/sdcflows/interfaces/reportlets.py", line 103, in _generate_report
abs(np.percentile(fmapdata[maskdata], 99.8)),
IndexError: boolean index did not match indexed array along dimension 1; dimension is 60 but corresponding boolean dimension is 112
Screenshots / relevant information:
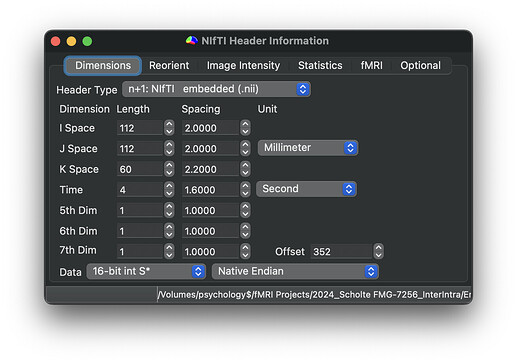
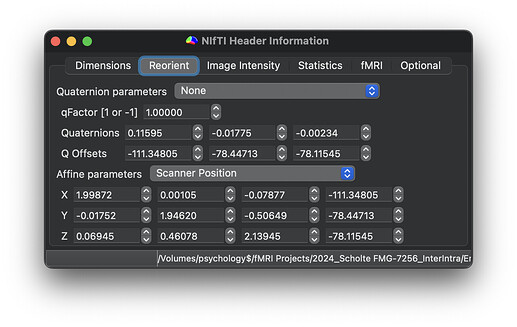
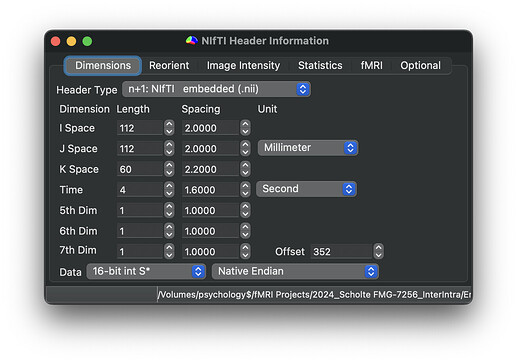
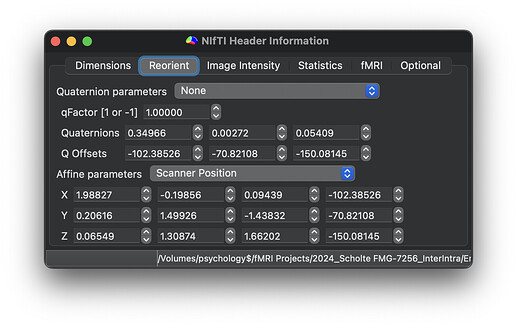
Here are the dimensions and orientation of one example TOP-UP scans:
Participant without error:
Participant with error: