Summary of what happened:
Hello,
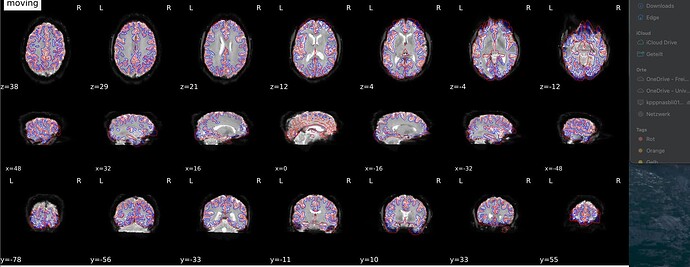
I am using fmriprep v23.0.2 and I am not sure if the registration of my functional images to the MNI space worked properly. When I look at the brain mask or the desc-preproc-bold.nii.gz file, they do not really look like being in the shape of the MNI template. For the T1, this looks fine.
So, I am not sure if this is normal, or if something went wrong somewhere?
When I compare the T1w and the functional image and/or brain mask, they all say being normalized to the MNI152NLin2009cAsym template, which would suggest to me that these should have the same shape.
I am also not sure what kind of mask I would now use for data analysis when I use this dataset for example with nilearn or conn.
Thank you for your help!
Best,
Klemens
Command used (and if a helper script was used, a link to the helper script or the command generated):
/opt/conda/bin/fmriprep /data /out participant --participant-label 01 02 03 04 05 06 07 08 09 10 --write-graph --me-output-echos --use-aroma --skull-strip-t1w force --skip-bids-validation --fs-license-file /freesurfer_license/freesurfer-license.txt --fd-spike-threshold 0.5 --verbose --nprocs 30 --omp-nthreads 4 -w /fmriprep_temp
Version:
fMRIprep V23.0.2
Environment (Docker, Singularity, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
Data in BIDS format. Validator showed no issues.