Summary of what happened:
I am using fMRIprep 23.1.3 to preprocess HCP-7T-rsfMRI data. However, I have encountered an issue during preprocessing where it gets stuck in the step “recon-all -parallel -subjid sub-100610 -sd /output/sourcedata/freesurfer -T2pial -cortribbon”. I have submitted 69 subjects via singularity-fMRIprep in HPC with 6 CPUs and 120GB memory. Unfortunately, this problem has occurred for 6 subjects. It seems that the “recon-all” step is taking significantly longer than expected to finish the “cortribbon” part of the preprocessing, while other normal preprocessing can fibish cortirbbon in 3 hours.
Command used (and if a helper script was used, a link to the helper script or the command generated):
#Run fmriprep
export SINGULARITYENV_TEMPLATEFLOW_HOME=$templateflow
unset PYTHONPATH;
singularity run --cleanenv \
-B $wd/$subj:/wd \
-B $bids:/bids \
-B $output:/output \
-B $fs_license:/fs_license \
-B $templateflow:$templateflow \
/ibmgpfs/cuizaixu_lab/xulongzhou/apps/singularity/fmriprep-23.1.3.simg \
/bids /output participant -w /wd \
--participant_label ${subj} \
--fs-license-file /fs_license/license.txt \
--n_cpus 12 --omp-nthreads 12 --mem-mb 120G \
--skip_bids_validation \
--task-id rest \
--output-spaces T1w:res-1 MNI152NLin2009cAsym:res-1 fsLR fsaverage \
--fmap-bspline \
--fmap-no-demean \
--force-syn \
--return-all-components \
--cifti-output 170k \
--stop-on-first-crash \
--fd-spike-threshold 0.2 \
--notrack
rm -rf $wd/$subj>
Version:
fMRIprep 23.1.3
Environment (Docker, Singularity, custom installation):
Both Singularity 3.7.0 and Docker
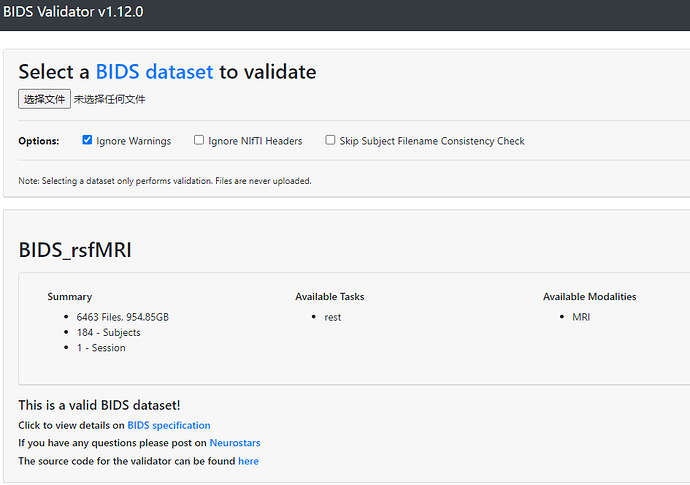
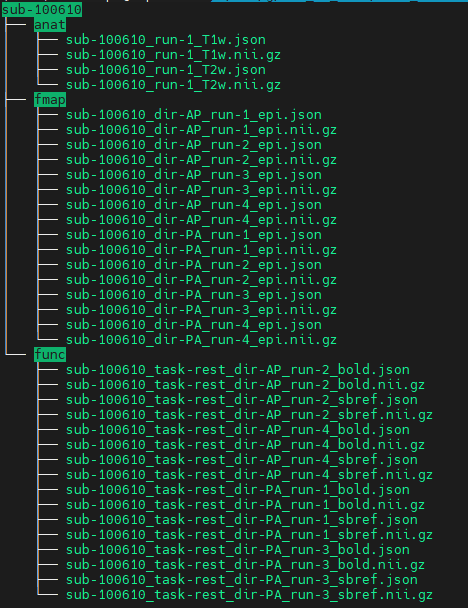
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS
Relevant log outputs (up to 20 lines):
230720-21:11:19,137 nipype.workflow INFO:
[Node] Finished "_autorecon_surfs1", elapsed time 0.113666s.
230720-21:11:20,876 nipype.workflow INFO:
[Node] Setting-up "fmriprep_23_1_wf.single_subject_146129_wf.anat_preproc_wf.surface_recon_wf.autorecon_resume_wf.cortribbon" in "/wd/fmriprep_23_1_wf/single_subject_146129_wf/anat_preproc_wf/surface_recon_wf/autorecon_resume_wf/cortribbon".
230720-21:11:20,901 nipype.workflow INFO:
[Node] Executing "cortribbon" <smriprep.interfaces.freesurfer.ReconAll>
230720-21:11:20,902 nipype.interface INFO:
resume recon-all : recon-all -parallel -subjid sub-146129 -sd /output/sourcedata/freesurfer -T2pial -cortribbon
230720-21:11:20,902 nipype.interface INFO:
resume recon-all : recon-all -parallel -subjid sub-146129 -sd /output/sourcedata/freesurfer -T2pial -cortribbon
230720-21:11:26,866 nipype.workflow INFO:
[Node] Setting-up "fmriprep_23_1_wf.single_subject_146129_wf.fmap_preproc_wf.wf_auto_00003.concat_blips" in "/wd/fmriprep_23_1_wf/single_subject_146129_wf/fmap_preproc_wf/wf_auto_00003/concat_blips".
230720-21:11:26,870 nipype.workflow INFO:
[Node] Executing "concat_blips" <niworkflows.interfaces.nibabel.MergeSeries>
230720-21:11:27,293 nipype.workflow INFO:
[Node] Finished "concat_blips", elapsed time 0.422901s.
230720-21:11:28,884 nipype.workflow INFO:
[Node] Setting-up "fmriprep_23_1_wf.single_subject_146129_wf.fmap_preproc_wf.wf_auto_00003.pad_blip_slices" in "/wd/fmriprep_23_1_wf/single_subject_146129_wf/fmap_preproc_wf/wf_auto_00003/pad_blip_slices".
230720-21:11:28,887 nipype.workflow INFO:
[Node] Executing "pad_blip_slices" <sdcflows.interfaces.utils.PadSlices>
230720-21:11:29,194 nipype.workflow INFO:
[Node] Finished "pad_blip_slices", elapsed time 0.306433s.