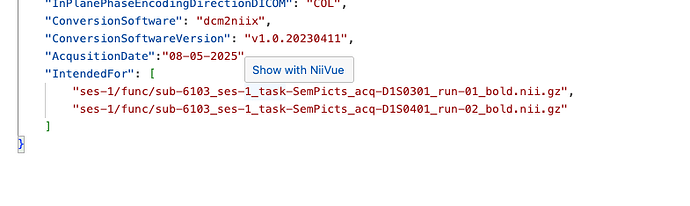
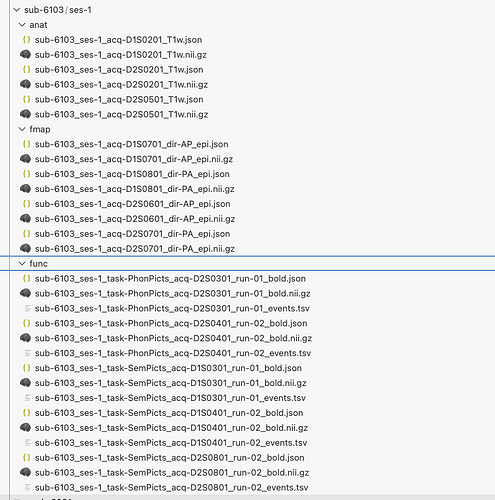
Thanks for helping out with this, I changed the fMRIprep to fmriprep/25.1.1 and used the dataset description as above, but still don’t get an output for susceptibility distortion correction ( * Susceptibility distortion correction: None appears in the final html report). It runs and generates the final report and all the files without producing any error. Content for sub-6081_ses-1_acq-D1S0501_dir-AP_epi.json is as follows:
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 127.75912,
"Manufacturer": "Philips",
"ManufacturersModelName": "Ingenia Elition X",
"InstitutionName": "VANDERBILT INSTITUTE OF IMAGING",
"InstitutionalDepartmentName": "MRI DEPT.",
"InstitutionAddress": "1161 21ST AVE. SOUTH",
"DeviceSerialNumber": "45051",
"StationName": "PHILIPS-8G70R22",
"BodyPartExamined": "BRAIN",
"PatientPosition": "HFS",
"SoftwareVersions": "5.7.1\\5.7.1.0",
"MRAcquisitionType": "2D",
"SeriesDescription": "TOPUP_APP",
"ProtocolName": "WIP TOPUP_APP",
"ScanningSequence": "SE",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"PulseSequenceName": "SEEPI",
"ImageType": ["ORIGINAL", "PRIMARY", "T2", "NONE"],
"SeriesNumber": 501,
"AcquisitionTime": "14:22:57.310000",
"AcquisitionNumber": 5,
"PhilipsRescaleSlope": 2.10525,
"PhilipsRescaleIntercept": 0,
"PhilipsScaleSlope": 0.0224536,
"UsePhilipsFloatNotDisplayScaling": 1,
"SliceThickness": 2.25,
"SpacingBetweenSlices": 2.25,
"EchoTime": 0.034229,
"RepetitionTime": 1.71597,
"MTState": false,
"FlipAngle": 90,
"CoilString": "MULTI COIL",
"PercentPhaseFOV": 100,
"PercentSampling": 100,
"EchoTrainLength": 47,
"PhaseEncodingSteps": 94,
"FrequencyEncodingSteps": 96,
"PhaseEncodingStepsOutOfPlane": 1,
"AcquisitionMatrixPE": 94,
"ReconMatrixPE": 96,
"ParallelReductionFactorInPlane": 2,
"ParallelAcquisitionTechnique": "SENSE",
"WaterFatShift": 12.8374,
"EffectiveEchoSpacing": 0.000304606,
"TotalReadoutTime": 0.0289376,
"AcquisitionDuration": 6.86387,
"PixelBandwidth": 2153.55,
"PhaseEncodingDirection": "j-",
"ImageOrientationPatientDICOM": [
0.992705,
-0.0919653,
-0.0779642,
0.101512,
0.986441,
0.128952 ],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20230411",
"AcqusitionDate":"07-10-2025",
"IntendedFor": [
"ses-1/func/sub-6081_ses-1_task-PhonPicts_acq-D1S0701_run-01_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-PhonPicts_acq-D1S0801_run-01_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-PhonPicts_acq-D1S0901_run-02_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-SemPicts_acq-D1S0301_run-01_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-SemPicts_acq-D1S0401_run-02_bold.nii.gz"
]
}
and content for sub-6081_ses-1_acq-D1S0601_dir-PA_epi.json is as follows
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"ImagingFrequency": 127.75912,
"Manufacturer": "Philips",
"ManufacturersModelName": "Ingenia Elition X",
"InstitutionName": "VANDERBILT INSTITUTE OF IMAGING",
"InstitutionalDepartmentName": "MRI DEPT.",
"InstitutionAddress": "1161 21ST AVE. SOUTH",
"DeviceSerialNumber": "45051",
"StationName": "PHILIPS-8G70R22",
"BodyPartExamined": "BRAIN",
"PatientPosition": "HFS",
"SoftwareVersions": "5.7.1\\5.7.1.0",
"MRAcquisitionType": "2D",
"SeriesDescription": "TOPUP_APA",
"ProtocolName": "WIP TOPUP_APA",
"ScanningSequence": "SE",
"SequenceVariant": "SK",
"ScanOptions": "FS",
"PulseSequenceName": "SEEPI",
"ImageType": ["ORIGINAL", "PRIMARY", "T2", "NONE"],
"SeriesNumber": 601,
"AcquisitionTime": "14:23:6.690000",
"AcquisitionNumber": 6,
"PhilipsRescaleSlope": 1.47643,
"PhilipsRescaleIntercept": 0,
"PhilipsScaleSlope": 0.0224536,
"UsePhilipsFloatNotDisplayScaling": 1,
"SliceThickness": 2.25,
"SpacingBetweenSlices": 2.25,
"EchoTime": 0.034229,
"RepetitionTime": 1.71879,
"MTState": false,
"FlipAngle": 90,
"CoilString": "MULTI COIL",
"PercentPhaseFOV": 100,
"PercentSampling": 100,
"EchoTrainLength": 47,
"PhaseEncodingSteps": 94,
"FrequencyEncodingSteps": 96,
"PhaseEncodingStepsOutOfPlane": 1,
"AcquisitionMatrixPE": 94,
"ReconMatrixPE": 96,
"ParallelReductionFactorInPlane": 2,
"ParallelAcquisitionTechnique": "SENSE",
"WaterFatShift": 12.835,
"EffectiveEchoSpacing": 0.00030455,
"TotalReadoutTime": 0.0289323,
"AcquisitionDuration": 6.87516,
"PixelBandwidth": 2153.55,
"PhaseEncodingDirection": "j",
"ImageOrientationPatientDICOM": [
0.992705,
-0.0919653,
-0.0779642,
0.101512,
0.986441,
0.128952 ],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20230411",
"AcqusitionDate":"07-10-2025",
"IntendedFor": [
"ses-1/func/sub-6081_ses-1_task-PhonPicts_acq-D1S0701_run-01_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-PhonPicts_acq-D1S0801_run-01_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-PhonPicts_acq-D1S0901_run-02_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-SemPicts_acq-D1S0301_run-01_bold.nii.gz",
"ses-1/func/sub-6081_ses-1_task-SemPicts_acq-D1S0401_run-02_bold.nii.gz"
]
}
Moreover, I am unable to append the json from BIDS validator here as it surpass the character limit. Is it possible to attach is as file.