Hello!
We successfully ran our first 7T subject through fmriprep; however, the anatomical segmentation appears to be failing in the temporal lobes. I’ve read through this thread which inquired about the manual editing option, but I’m wondering if there are tags we can add to our command line to help with this segmentation (similar to how FSL’s BET allows for -f and -g to adjust the estimation of the brain). This does not appear to be a brain mask issue, but rather a segmentation issue; it also seems a bit much for manually editing.
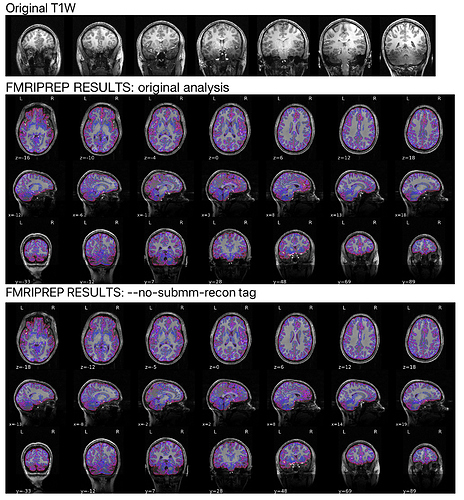
For reference, the red outlines are the lh.pial and rh.pial and the yellow are the lh.white and rh.white segmentations from the surf directory…overlaid on the T1/brainmask.
What is your exact command line? And more specifically, did you use the --no-submm-recon flag?
Hello!
I used:
fmriprep-docker /path/to/data/dir /path/to/output/dir participant -v
We didn’t use any other tags.
I’ll give the --no-submm-recon a try!
Thanks for the suggestion!
Hi, the --no-submm-recon option should be even worse (by theory). Could you share one of those T1w so that we can see whether the inhomogeneity correction can be made more robust?
Sure thing. I just started re-running it, but as soon as it completes I’ll submit images of the original T1w, the first pass through fmriprep w/ no tags, and then the no-submm tag.
I’m working on this improvement - https://github.com/poldracklab/niworkflows/pull/342 which may improve INU correction on your data. I’ll let you know when I have some docker image you can check.
Hi @jrobinson,
I have uploaded a new docker image that may help fix this issue. If fmriprep-docker was working for you, this command should also run:
docker run --rm -it -v /path/to/data/dir:/data:ro -v /path/to/output/dir:/output \
poldracklab/smriprep:n4-improvements /data /output participant \
--participant-label <participant-id> -vv
where <participant-id> would be that of the example shown in the first post.
Looking forward to your feedback
Thanks so much for your help so far. Here’s what we have:
When I try to run your updated command, I run into the following error:
Status: Downloaded newer image for poldracklab/smriprep:n4-improvements
docker: Error response from daemon: Failed to inspect container 0590bae6eed1760348ef653f8c5da34a8c0b0cc309e3b357583390459d7a39c6: Error response from daemon: readlink /var/lib/docker/overlay2/l: invalid argument.
ERRO[0227] error waiting for container: context canceled
Any suggestions on how to correct this?
Best,
Jenny
Seems like a problem with your docker installation, not smriprep. Are you on a Mac system?
If so:
$ screen ~/Library/Containers/com.docker.docker/Data/com.docker.driver.amd64-linux/tty
$ umount /var/lib/docker/overlay2
$ rm -rf /var/lib/docker
Should clean up the Docker installation (beware that any previously cached images will be deleted and will need to download again).
Please let me know whether you are using Linux or Windows and I’ll try to find corresponding solutions.
Hi!
Thanks for the feedback. My latest and greatest issue is that I keep getting this error:
Traceback (most recent call last):
- File “/usr/local/miniconda/bin/smriprep”, line 11, in *
- load_entry_point(‘smriprep==0.1.1+17.g5779bbf’, ‘console_scripts’, ‘smriprep’)()*
- File “/usr/local/miniconda/lib/python3.7/site-packages/smriprep/cli/run.py”, line 14, in main*
- return build_opts(opts)*
- File “/usr/local/miniconda/lib/python3.7/site-packages/smriprep/cli/run.py”, line 190, in build_opts*
- 'ERROR: a valid license file is required for FreeSurfer to run. '*
RuntimeError: ERROR: a valid license file is required for FreeSurfer to run. sMRIPrep looked for an existing license file at several paths, in this order: 1) command line argument --fs-license-file; 2) $FS_LICENSE environment variable; and 3) the $FREESURFER_HOME/license.txt path. Get it (for free) by registering at https://surfer.nmr.mgh.harvard.edu/registration.html
…which is puzzling because the license file is an environment variable; and even if I use the --fs-license-file tag with the appropriate path to the license, it’s still not happy…yet, if I run fmriprep-docker, it runs fine without any fits.
Hi @jrobinson, fmriprep-docker will look for your license file, using the FS_LICENSE variable if it’s defined, and make it available inside your Docker container. You’ll need to do the same with sMRIPrep. If you have the FS_LICENSE environment variable, you can add the following: to your Docker command (before the image):
-v $FS_LICENSE:/opt/freesurfer/license.txt:ro
More generally, to modify commands, you can use fmriprep-docker to get the full Docker command line. Whenever you run it, you should see a line starting with RUNNING:. That is what is actually passed to Docker, and if you look in there, you should see something similar to the above that you can use.
1 Like
Sorry I failed to reply to this last week.
My apologies for the delayed response. I really appreciate your help, @effigies & @oesteban! It’s currently running, so hopefully I’ll be able to report back soon with the results 
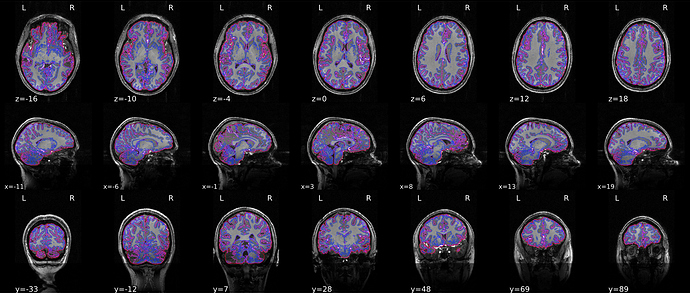
Here’s the result of running the new smriprep:
Unfortunately, we’re still getting poor segmentation in the anterior temporal lobe.
Hi @jrobinson, any success with using FreeSurfer directly? For consistency, we pass in the ANTs-extracted brain mask, but if you run FreeSurfer separately, it will use its own skull-stripping, which may work better in some cases. I would give that a try, just to see.
I would also suggest the ANTs discussion site, to see if they can help you find a set of parameters that works for you. It’s possible that we can incorporate them.
I think we may be coming to a point, though, where it’s going to be necessary to allow users to pass in a brain mask. I’m not sure how plausible it’s going to be to tweak the ANTs skullstripping algorithm to handle all of the edge cases while maintaining good performance for the majority of images.
Hi @jrobinson,
I just picked out some detail from your caption that is bugging me.
There is a clear cut in brightness under the nose. That makes me think that we are not talking of just one T1w image and that you might have a large number of them. Is that correct? Why is that sharp change in contrast below the cerebellum?
That could be confusing the atlas-based brain extraction of antsBrainExtraction.
I’ll give FreeSurfer a shot!
@oesteban, we do have a number of subjects (~40). I see what you’re referring to, and I’ll check the other scans to see if it’s just this subject or an issue with the sequence/headcoil. At any rate, I’ll run a few others and report back, along with the FreeSurfer results.
I meant that it seems you have several T1w images per subject. In such scenario, fMRIPrep would realign them and create a t1w template fusing them.
Can you run find /path/to/bids/folder -maxdepth 4 -name "*_T1w.nii.gz"?