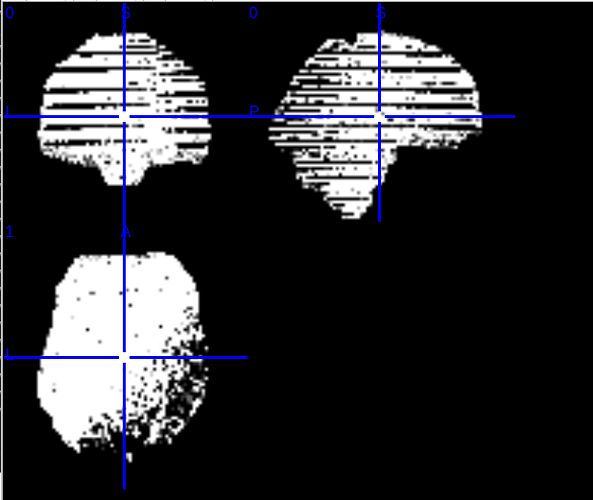
Summary of what happened: I ran fMRIprep on multi-echo data preprocessed with fmriprep using the flag –me-output-echos and then processed the output echos (in native space) with tedana. The tedana output (every image) has an odd, stripey drop-out and I can’t seem to understand why. Screenshot is of the adaptive mask, and all post-tedana subject images have the same drop-out pattern. Any ideas of why this might be happening? Thanks in advance for your guidance!!
Command used (and if a helper script was used, a link to the helper script or the command generated):
>tedana -d ./fmri_preproc/derivatives/fmriprep_out/sub-pilot2decob/func/sub-pilot2decob_task-decobi_run-1_
echo-1_desc-preproc_bold.nii.gz ./fmri_preproc/derivatives/fmriprep_out/sub-pilot2decob/func/sub-pilot2de
cob_task-decobi_run-1_echo-2_desc-preproc_bold.nii.gz ./fmri_preproc/derivatives/fmriprep_out/sub-pilot2d
ecob/func/sub-pilot2decob_task-decobi_run-1_echo-3_desc-preproc_bold.nii.gz ./fmri_preproc/derivatives/fm
riprep_out/sub-pilot2decob/func/sub-pilot2decob_task-decobi_run-1_echo-4_desc-preproc_bold.nii.gz ./fmri_preproc/derivatives/fmriprep_out/sub-pilot2decob/func/sub-pilot2decob_task-decobi_run-1_echo-5_desc-preproc_bold.nii.gz -e 12.200 28.920 45.64
0 62.360 79.080 --prefix sub-pilot2decob_task-decobi_run-1_desc-tedana --mask ./fmri_preproc/derivatives/
fmriprep_out/sub-pilot2decob/func/sub-pilot2decob_task-decobi_run-1_desc-brain_mask.nii.gz --masktype none --tedpca kundu --out-dir ./fmri_preproc/derivatives/tedana/sub-pilot2decob/func/sub-pilot2decob_task-decobi_run-1 --verbose
Version:
tedana 25.0.1
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS